Fig. 4.
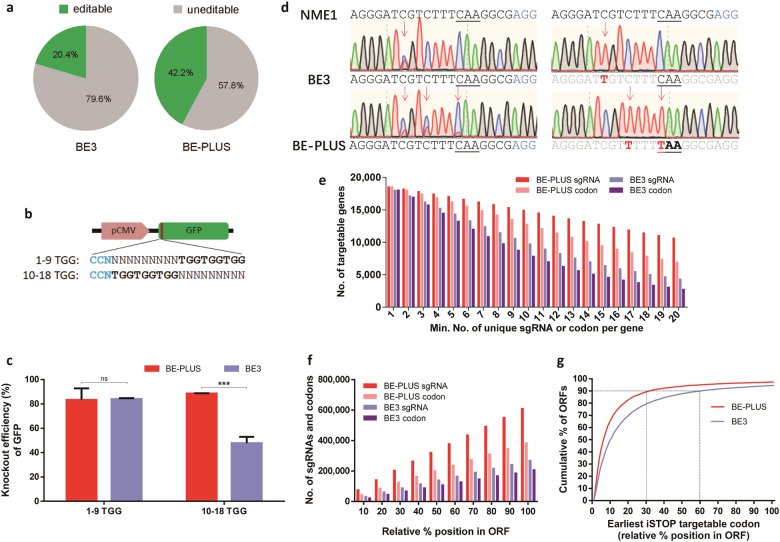
BE-PLUS is more efficient in inducing stop codons. a Cs editable by BE3 and BE-PLUS in the human genome were predicted based on the editing window of BE3 and BE-PLUS, respectively (all Cs and Gs in the chromosomes of the human reference genome hg38 were counted). Editable Cs are shown in green. b Scheme of GFP-iSTOP reporter system with 3 iSTOP codons (TGG) at positions 1–9 or 10–18 (from the distal side of the PAM). c Analysis of the GFP shutdown efficiency of BE-PLUS and BE3 using the GFP-iSTOP reporter system. The bar graph shows the GFP shutdown efficiency of BE-PLUS or BE3 when 1–9 or 10–18 TGG reporter vectors were used. GFP shutdown efficiency was calculated as the percentage of GFP knockout cells among the GFP-positive cells in the control group. Error bars (±) indicate the standard deviations of 3 replicates. ns, not significant, P ≥ 0.05; ***P < 0.001. d The plasmids expressing NME1 sgRNA (Supplementary information, Table S8) was co-transfected with either BE3 or BE-PLUS into HEK293FT cells. PCR products around the sgRNA binding site were subjected to Sanger sequencing (left), and T-A cloning and sequencing (right). e Statistical analysis of unique iSTOP sgRNAs or codons for BE3 and BE-PLUS in the entire human transcriptome. The unique iSTOP sgRNAs were identified over the genome dependent on the base editing window of BE-PLUS (from positions 4 to 16) or BE3 (from positions 4 to 8). The number of genes in the human genome that contain at least one unique iSTOP codon or are targeted by at least one iSTOP sgRNA for BE3 or BE-PLUS were calculated. f Statistical analysis of the iSTOP codons or sgRNAs for BE-PLUS or BE3. The bars represent the cumulative number of codons or sgRNAs for BE-PLUS or BE3. g The distribution of iSTOP codons over the targetable ORFs for BE-PLUS or BE3. The percentage of ORFs that can be disrupted and the relative positions of the iSTOP codons are shown