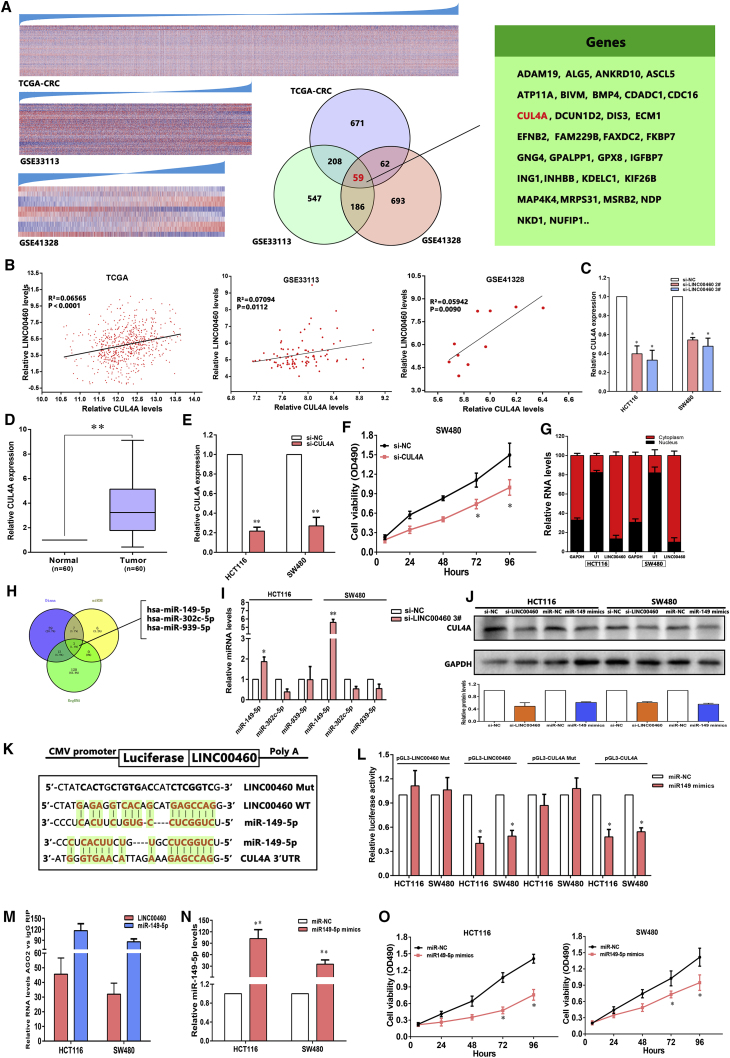
Figure 6.
LINC00460 Represses CUL4A Expression by Competing for miR-149-5p in Cytoplasm, Thus Facilitating CRC Cell Proliferation
(A) Heatmap of protein-coding genes that were significantly and positively co-expressed with LINC00460. The genes were arranged from top to bottom in ascending order of their correlation with LINC00460. Venn diagrams of LINC00460-associated genes among TCGA-CRC, GSE33113, and GSE41328 are shown. (B) The relationship between LINC00460 expression and CUL4A mRNA levels was analyzed in the profile of CRC patient tissue from TCGA and GEO. (C) The levels of CUL4A mRNA expression were determined by real-time qPCR when HCT116 and SW480 cells were transfected with si-NC and si-LINC00460. (D) By real-time qPCR assays, the level of CUL4A was remarkably upregulated in 60 pairs of CRC tissues. (E and F) Cell viability was determined upon knockdown of CUL4A in SW480 cells. (E) HCT116 and SW480 cells were transfected with si-CUL4A. (F) MTT assays were used to determine the cell viability for si-CUL4A-transfected SW480 cells. (G) Relative LINC00460 levels in HCT116 and SW480 cell cytoplasm or nucleus were detected by real-time qPCR. GAPDH was used as cytoplasm control, and U1 was used as nuclear control. (H) Potential binding sites of LINC00460-miRNA-CUL4A using the online software programs are shown. (I) Real-time qPCR assays detected the expression of miR-149-5p/miR-302c-5p/miR-939-5p after knockdown of LINC00460 in HCT116 and SW480 cells. (J) Western blot detection of the CUL4A protein levels in HCT116 and SW480 cells after knockdown of LINC00460 and overexpression of miR-149-5p is shown. (K) A photograph showing the predicted interaction between LINC00460/miR-149-5p and miR-149-5p/CUL4A mRNA through complementary base pairs is shown. (L) Both LINC00460 and CUL4A are targeted by miR-149-5p. The 3′ UTR of CUL4A mRNA and full length of LINC00460 were respectively inserted downstream of a luciferase gene. The reporter vector was co-transfected with a Renilla luciferase vector (for normalization) to SW480 and HT116, which were treated by miR-149-5p mimics or control mimics. The luciferase signals of both reporter genes were significantly decreased when cells were treated with miR-149-5p mimics. Mutant 3′ UTR of CUL4A and LINC00460 is not affected by miR-149-5p. (M) LINC00460 and miR-149-5p RNA levels in immunoprecipitates are presented as fold enrichment in AGO2 relative to IgG immunoprecipitates. (N) miR-149-5p levels were examined in HCT116 and SW480 cells after transfection with mimics. (O) MTT assays showed that cell proliferation was suppressed by miR-149-5p overexpression. The data are presented as the mean ± SD of three independent experiments; *p < 0.05; **p < 0.01.