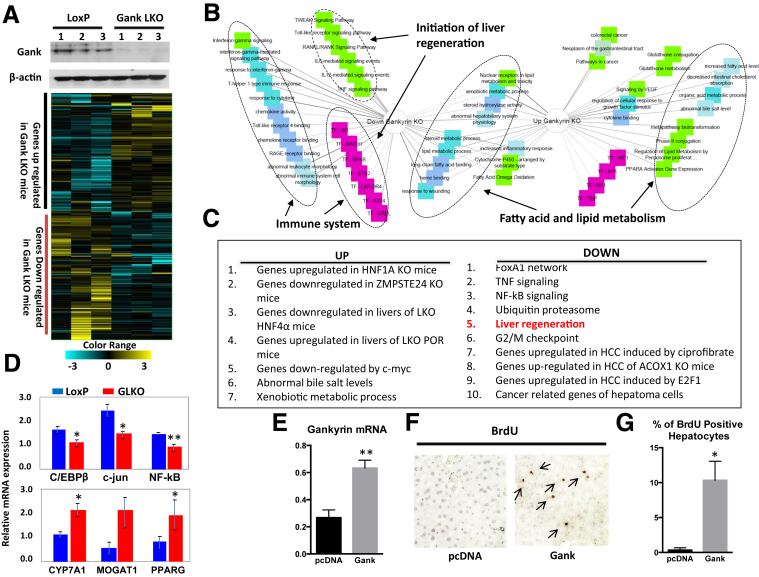
Figure 2.
Examination of signaling pathways that are altered in livers of Gank LKO mice. (A) RNA sequencing–based heat map of genes with altered expression in livers of Gank LKO mice. Three animals of each genotype were used. Upper image shows deletion of Gank by Western blot in the livers of mice used for RNA sequencing. (B) Schematic presentation of networks that are changed in livers of Gank LKO mice. (C) List of main pathways altered in Gank LKO mice. (D) An example of confirmation of genes overexpressed in livers of Gank LKO mice using quantitative RT-PCR with 3 mice per genotype. Two technical replicates were performed for each biological sample. Upper: mRNA levels for genes involved in liver regeneration. Bottom: Levels of mRNAs that are increased in GLKO mice. P values were as follows: C/EBPβ (P < .04); c-jun (P < .02); nuclear factor-κB (NF-κB) (P < .002), CYP7A1 (P < .04), MOGAT1 (P > .08), and PPARG (P < .04). (E) Levels of Gank mRNA in the livers transfected with empty vector plasmid DNA vector and with vector expressing Gank (P < .0015). (F) Immunostaining of the livers with antibodies to BrdU. Arrows show BrdU-positive hepatocytes. (G) Bar graphs show the percentage of BrdU-positive hepatocyte in livers transfected with empty vector and with plasmid expressing Gank (P < .02). Three mice were used for the empty vector and 3 mice were used for the plasmid expressing Gank experiment, with 3 fields analyzed per mouse. An unpaired Student t test. ∗P < .05, ∗∗P < .01 LoxP vs GKO.