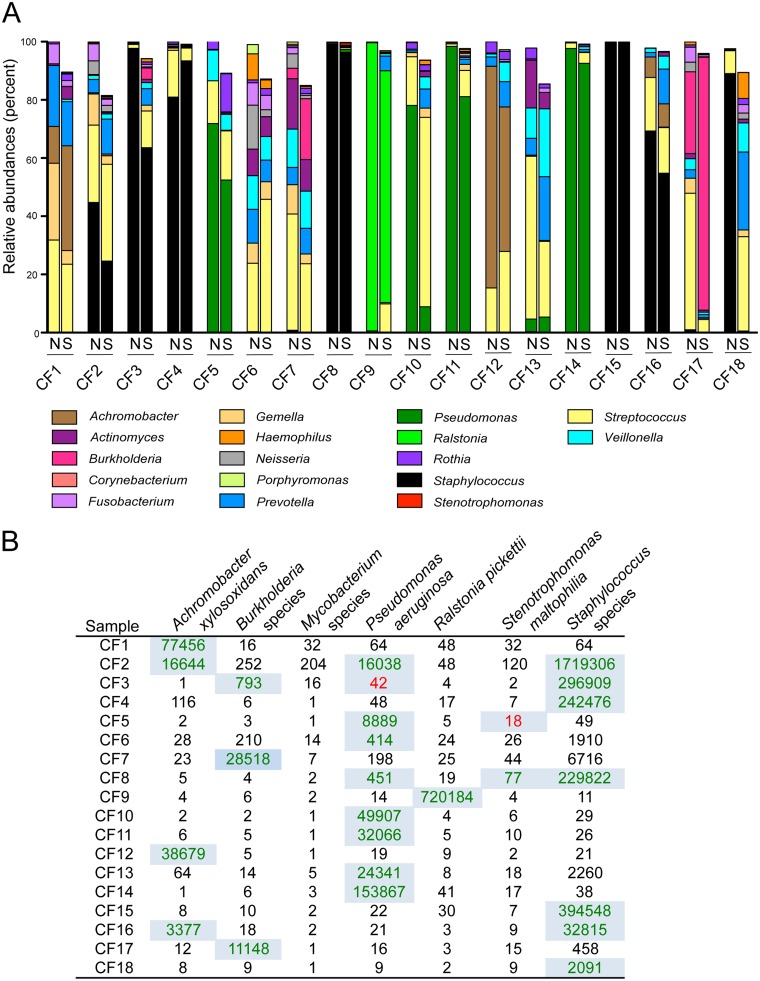
FIG 4 .
Bacterial community profiles detected by NanoString and 16S rRNA gene deep sequencing (MiSeq) methods correlate and are confirmed by culture. (A) Eighteen CF sputum samples, labeled with the subject number (CF1 to -18), were analyzed by both NanoString (N) and 16S rRNA gene deep sequencing (MiSeq [S]). The percentages of relative abundance of taxa detected by both methods are shown. Only the taxa measured in our NanoString code set are included in the deep sequencing profiles, but relative abundance was determined as a percentage of total reads, not only reads represented in the NanoString code set (data in Table S4). In cases where NanoString probes detected different species within the same genus, counts from individual species-specific probes were summed to correspond with the genus-level assignments for the MiSeq data. (B) The counts for NanoString probe sets correlating with CF pathogens detected by culture are shown. See Table S3D for data for all genera analyzed. Highlighted numbers indicate that the corresponding pathogen was also detected by clinical culture analysis. The numbers in red in CF3 and CF5 were reported as being “rare” in the clinical microbiology analysis and were at very low levels (<0.5%) in the MiSeq-based community profiles for those samples. For CF sputum specimens, the clinical lab reports staphylococci only if they are identified as S. aureus.