Figure 6.
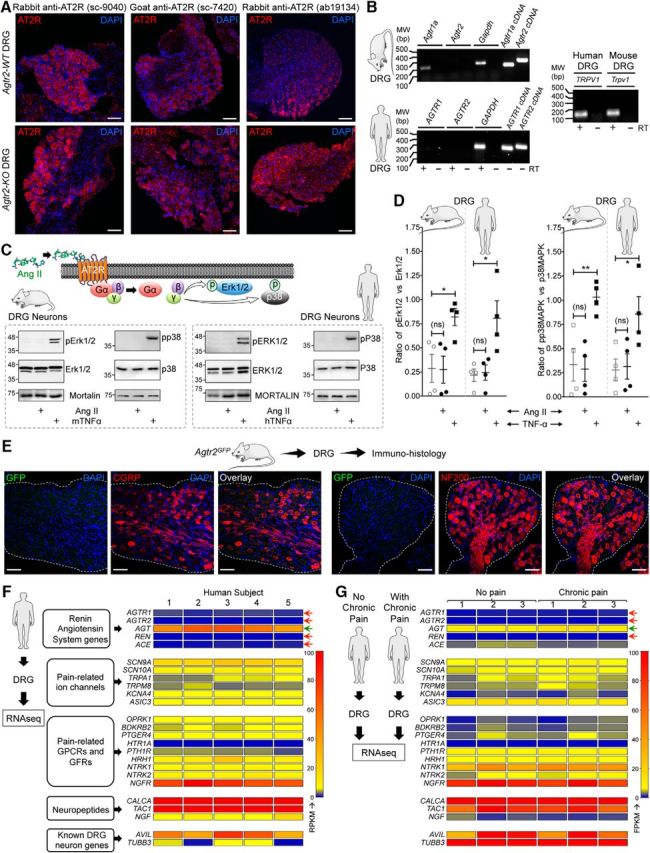
AT2R expression is not detected on mouse and human DRG sensory neurons. A, Representative confocal microscopy images of DRG tissue sections from FVB-Agtr2-WT and FVB-Agtr2-KO mice immunostained with routinely used anti-AT2R antibodies (red). Scale bar, 50 μm. No difference in AT2R signal intensity can be seen in the DRG sections from both mouse genotypes. B, Representative agarose gel electrophoresis images of RT-PCR amplification of AT1R and AT2R genes (Agtr1 and Agtr2, respectively) from the total RNA isolated from mouse and human DRGs. Plasmids containing mouse and human AT1R and AT2R cDNAs are used as a positive control. In addition, TRPV1 amplification is used as a positive control for DRG tissue. Numbers on the left denote DNA molecular weight markers (in base pairs). C, Ang II (1 μm; 30 min) does not induce phosphorylation of ERK1/2 and p38 MAPK, indicative of Ang II/AT2R activation, in cultured mouse and human DRG neurons. TNF-α (10 nm) is used as a positive control for induction of ERK1/2 and p38 MAPK phosphorylation. D, Quantification of the extent of ERK1/2 and p38 MAPK phosphorylation levels in mouse and human DRG neurons in response to Ang II exposure (1 μm; 30 min), as shown in representative immunoblots in Figure 3A. TNF-α (10 nm; 30 min) is used as a positive control for induction of ERK1/2 and p38 MAPK phosphorylation. Data are presented as individual experimental replicates, with mean ± SEM marked therein (n = 4 per group). *p < 0.05, **p < 0.01, and not significant (ns) versus respective comparison groups, one-way ANOVA with Tukey's multiple-comparisons post hoc test. E, The AT2R gene (Agtr2) is not expressed in neurons and non-neuronal cells in mouse DRG, as verified by lack of GFP signal (as a surrogate marker) in DRG sections (L2–L5) from Agtr2GFP reporter mice in which the Agtr2 promoter drives GFP expression. Representative confocal microscopy images of DRG sections stained with anti-GFP antibodies, along with CGRP and NF200 antibodies to mark peptidergic and myelinated sensory neurons. DAPI was used as a nuclear stain. Scale bar, 50 μm. F, G, Heat map showing mRNA expression levels (from RNAseq experiments) of RAS genes compared with critical pain-associated genes in human DRG tissue (F). No alteration in the mRNA levels of RAS genes can be observed in DRGs obtained from humans without or with chronic pain conditions (G). Red arrows indicate no reliable mRNA expression levels and green arrow indicates considerable mRNA expression of RAS genes.
