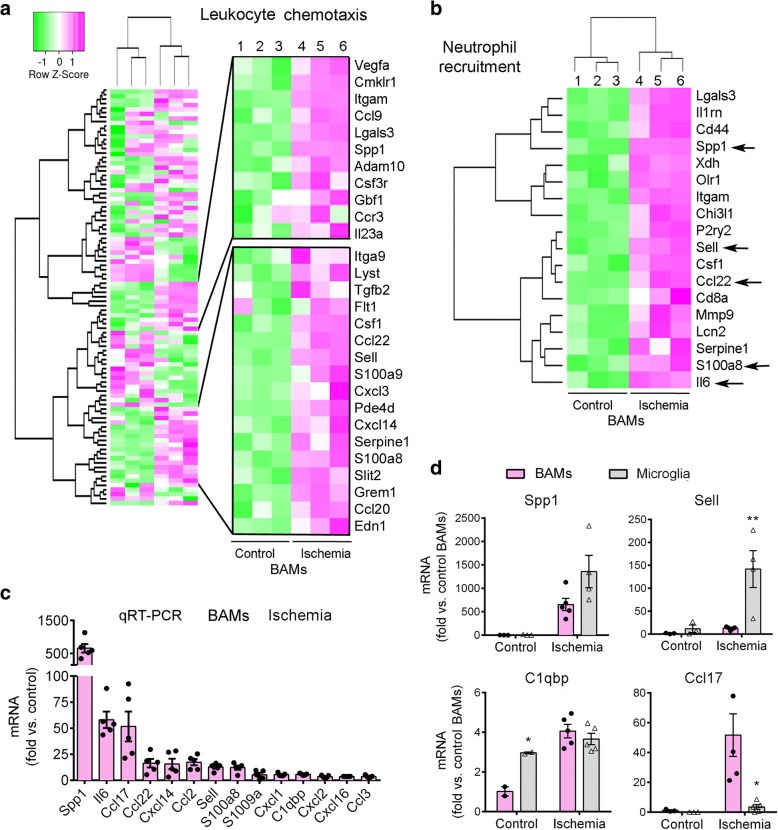
Fig. 3.
CD163+ BAMs upregulate the expression of leukocyte chemoattractants after ischemia. a) Heatmap showing the profile expression of upregulated (pink) and downregulated (green) genes in the GO term Leukocyte chemotaxis. A set of genes upregulated after ischemia is highlighted. b) Heatmap of genes of the IPA category Recruitment of neutrophils. Some of these genes (arrows) were later analyzed by qRT-PCR in RNA of cells sorted from a different set of rats. c) In a different group of controls (n = 3) and ischemic rats (n = 5) (16 h of reperfusion), we sorted CD163+ BAMs, extracted RNA and carried out qRT-PCR to validate up-regulation of some of the genes previously identified in the microarray. Values are expressed as fold vs. the mean value of control CD163+ cells and are shown as the mean±SD. d) Gene expression as assessed by qRT-PCR in sorted CD163+ BAMs and sorted microglia (n = 3 controls and 5 ischemic rats per each cell type). All values are normalized versus the mean value of controls and are represented as the mean±SD. Generally, induction of gene expression after ischemia is similar or lower in BAMs than microglia, with the exception of some genes like Ccl17 that was induced after ischemia in BAMs but not microglia. Two-way ANOVA followed by the Bonferroni test, *p < 0.05 and **p < 0.01 microglia vs. BAMs