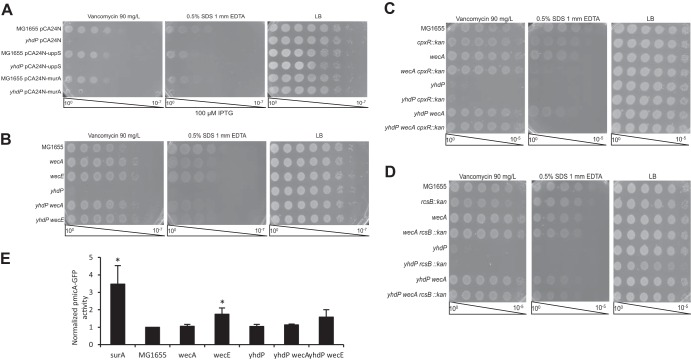
FIG 3 .
Suppression does not relate to Und-P availability or stress responses. (A) To determine whether the ΔyhdP strain is suppressed by relieving Und-P stress, EOPs were performed on strains carrying the indicated overexpression constructs. Overexpression of uppS and murA did not suppress the ΔyhdP strain, demonstrating that the ΔyhdP strain's phenotypes are not caused by effects of Und-P stress on peptidoglycan. (B) EOPs were performed to determine whether disruptions in ECA biosynthesis that increased Und-P availability for peptidoglycan synthesis (ΔwecA) and that decreased Und-P availability for peptidoglycan synthesis (ΔwecE) both suppress the ΔyhdP strain phenotypes. Both of these mutations suppressed the ΔyhdP strain phenotypes to an equal extent, suggesting that suppression is unrelated to Und-P availability. (C) EOPs were performed to determine whether the Cpx response was responsible for suppression of the ΔyhdP strain's phenotypes by disruptions of ECA biosynthesis. Suppression was observed with wecA deletion, even in the presence of cpxR deletion, demonstrating that the Cpx response is not necessary for suppression. (D) EOPs were performed to determine whether the Rcs response was required for suppression. Suppression was observed in the presence of rcsB deletion, demonstrating that the Rcs response is not necessary for the suppression of the ΔyhdP strain's phenotypes. EOPs images are representative of three independent experiments. (E) Activity of a σE reporter was assayed to determine whether suppression of the ΔyhdP strain correlated with σE activation. Suppression of the ΔyhdP strain's phenotypes did not correlate with σE activation, suggesting this is not the mechanism of suppression. Data shown are the average results of three independent biological replicates ± the SEM. Significance was calculated using the Mann-Whitney test. *, P < 0.05 compared to the appropriate parent strain (MG1655 or ΔyhdP).