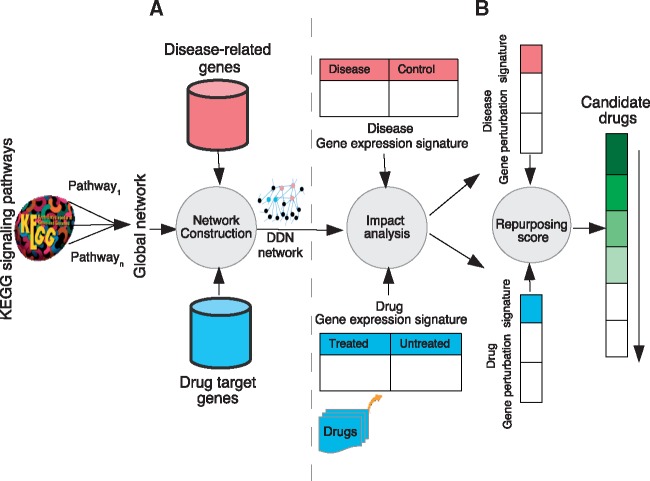
Fig. 1.
Framework overview. (A) We construct a global network (GN) that is the union of all KEGG human signaling pathways. For each drug-disease pair, we extract a subgraph of GN, namely DDN, consisting of all shortest paths between two sets of disease-related genes and drug targets. (B) We then generate gene perturbation signatures of drug-disease pairs by applying a system-level analysis on their gene expression signatures in the drug-disease network (DDN). A comparative analysis is applied on drug and disease gene perturbation signatures. A repurposing score is assigned to each drug-disease pair. Finally, a ranked list of drugs with potential therapeutic effects for the given disease is generated based on repurposing scores