Fig. 4.
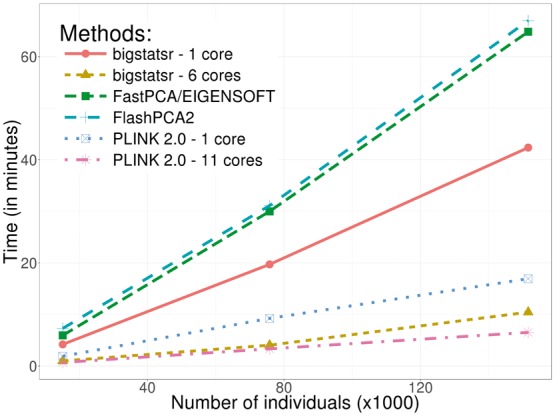
Benchmark comparisons between randomized partial singular value decomposition available in FlashPCA2, FastPCA (fast mode of SmartPCA/EIGENSOFT), PLINK 2.0 (approx mode) and package bigstatsr. It shows the computation time in minutes as a function of the number of samples. The first 10 principal components have been computed based on the 93 083 SNPs which remained after thinning
