Fig. 3.
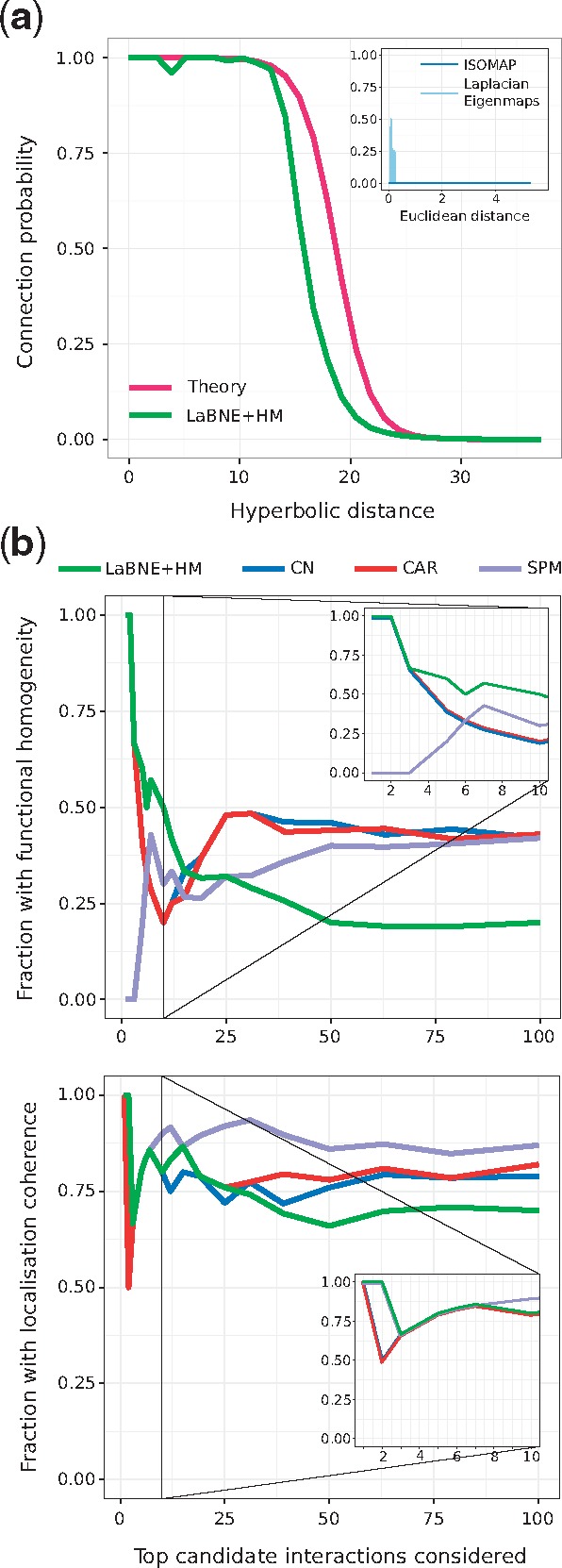
(a) Connection probability as a function of the hyperbolic or Euclidean (inset) separation between protein pairs. The probabilities predicted by the PSM (Theory) and the ones obtained by mapping the network to a geometric space with LaBNE + HM, ISOMAP and Laplacian Eigenmaps are shown. (b) We compared the top-100 disconnected proteins that are closest to each other in (LaBNE + HM) with candidate protein interactions from representative link predictors of different classes (see Supplementary Fig. S8 for the complete analysis). The plot shows how the fraction of potential interactions with functional homogeneity and localization coherence changes as more protein pairs are assessed. Insets focus on the top-10 candidate pairs. CN: Common Neighbours, CAR: Cannistraci-Alanis-Ravasi index, SPM: Structural Perturbation Method
