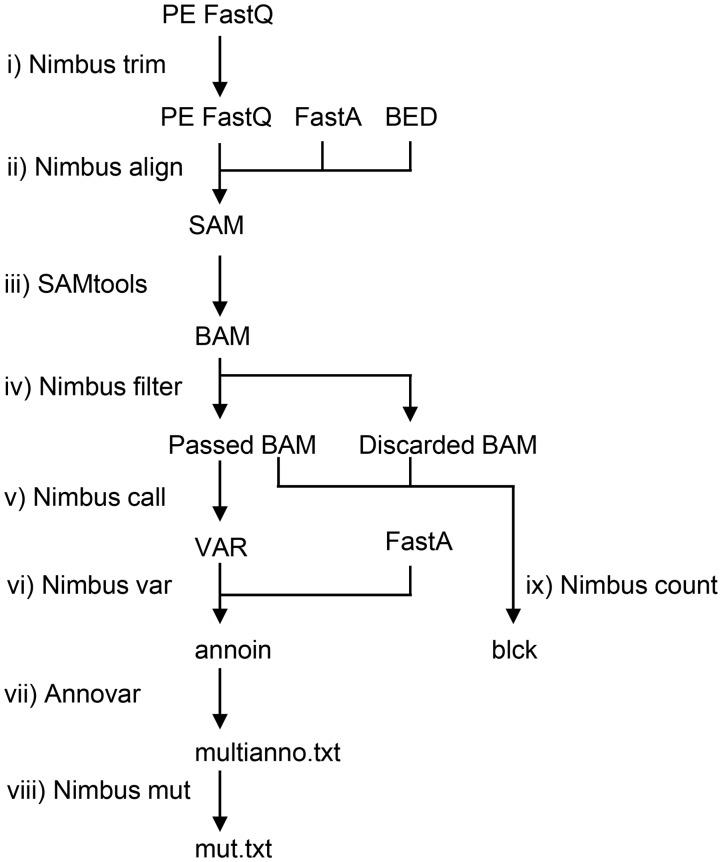
Fig. 1.
Nimbus analysis workflow. (i) Reads are first trimmed using Nimbus trim and (ii) subsequently aligned to the reference design using Nimbus align. (iii) The resulting SAM file is sorted and converted to BAM format using SAMtools. (iv) The alignments with four or more differences are copied to the discarded BAM files. Alignments with fewer than four differences are written to the passed BAM files that are used in the downstream analysis. The threshold for filtering can be adjusted. (v) Nimbus call records all differences between the reference sequence and the (passed) alignments in a custom var format. (vi) From these var files, variants are distilled by Nimbus var and reported in the tab-delimited ANNOVAR input format. (vii) The variants files are annotated with ANNOVAR (13) and (viii) converted to the mut.txt format which is readable by the IGV (14). (ix) In parallel with the variant calling, the read depths per amplicon are determined by Nimbus count from both the passed and discarded alignments. This information is recorded in blck files which can be used to determine amplicon performance