Figure 1.
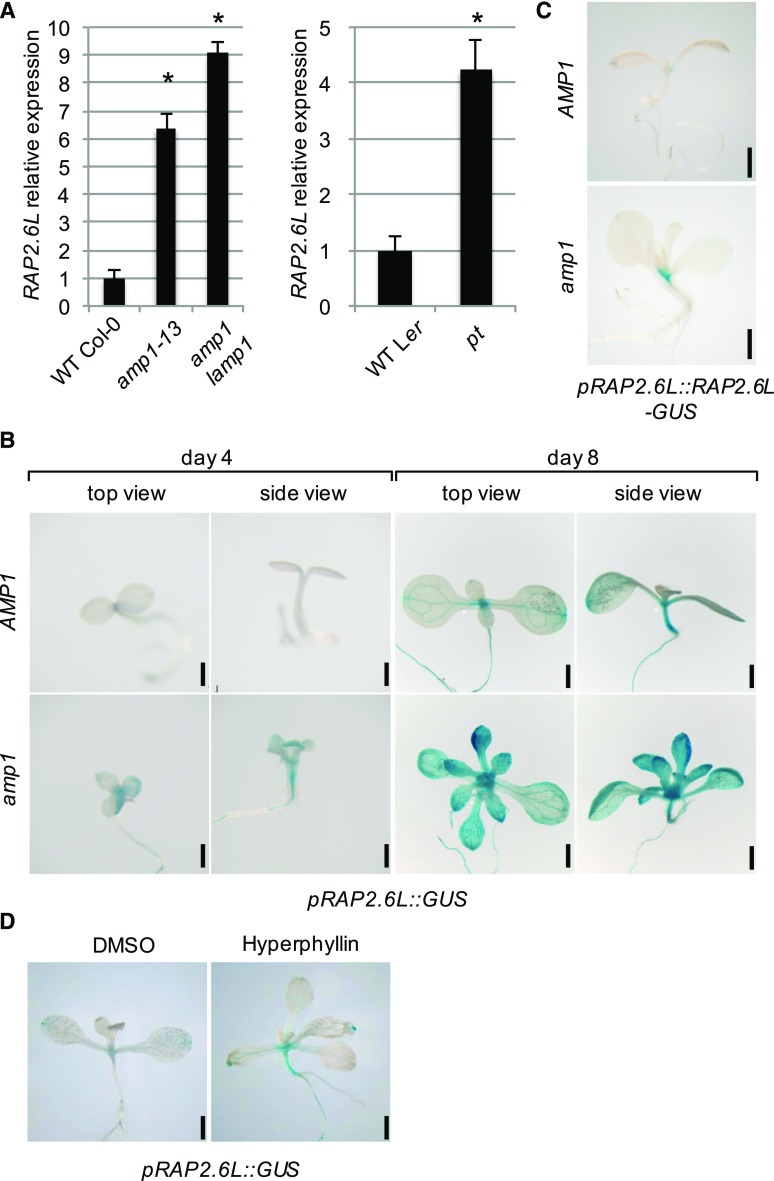
RAP2.6L expression is upregulated in amp1. A, qPCR analysis of RAP2.6L expression in 8-d-old seedlings of the indicated lines. Fold changes compared to the respective wild-type (WT) are shown. Error bars indicate the SE calculated from three biological replicates (at least 30 seedlings per replicate; replicates were grown at the same time on different petri dishes) after normalization to UBIQUITIN-CONJUGATING ENZYME (UBC). Asterisks indicate a significant difference (Student’s two-tailed t test; P < 0.05). B, pRAP2.6L::GUS activity in wild-type Col-0 (WT) and the amp1-1 mutant at 4 and 8 DAG. C, pRAP2.6L::RAP2.6L-GUS activity in wild-type Col-0 (WT) and the amp1-1 mutant at 8 DAG. D, pRAP2.6L::GUS activity in 10-d-old wild-type Col-0 seedlings grown in liquid medium containing either 0.5% DMSO (mock) or 30 μm hyperphyllin. Bars = 1 mm (B–D).