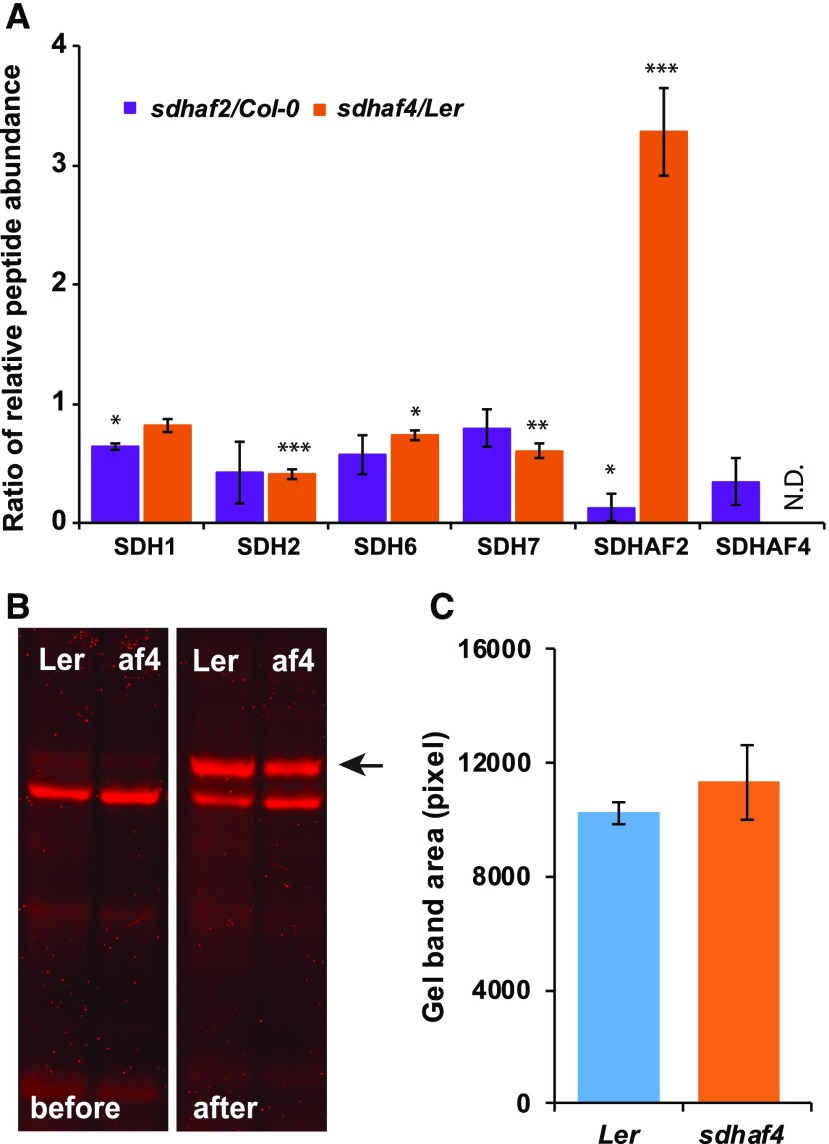
Figure 4.
Abundances of SDH2 and SDHAF2, but not FAD-bound SDH1, are altered in sdhaf4. A, MRM was used to detect peptides from SDH subunits and assembly factors. Shown are the ratios of peptides (mutant to wild type) based on whole mitochondria protein samples (50 µg; n = 3). N.D. indicates not detected in sdhaf4. Error bars indicate se. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (Student’s t test for sdhaf2:Col-0 or sdhaf4:Ler). B, A FAD-bound protein assay was performed to compare FAD binding with SDH1 in sdhaf4 (af4) and Ler. Mitochondrial proteins (10 µg) were separated by SDS-PAGE, following a gel incubation in 10% (v/v) acetic acid for 30 min. FAD fluorescence scans were performed before and after the acetic acid treatment using Typhoon Trio Laser (Amersham Biosciences) and filters Cy5 (670 bp) and Cy3 (580 bp). The FAD-bound SDH1 band became visible after acetic acid incubation (marked with a black arrow). C, Quantification of FAD-bound SDH1 bands on gels, with three replicates.