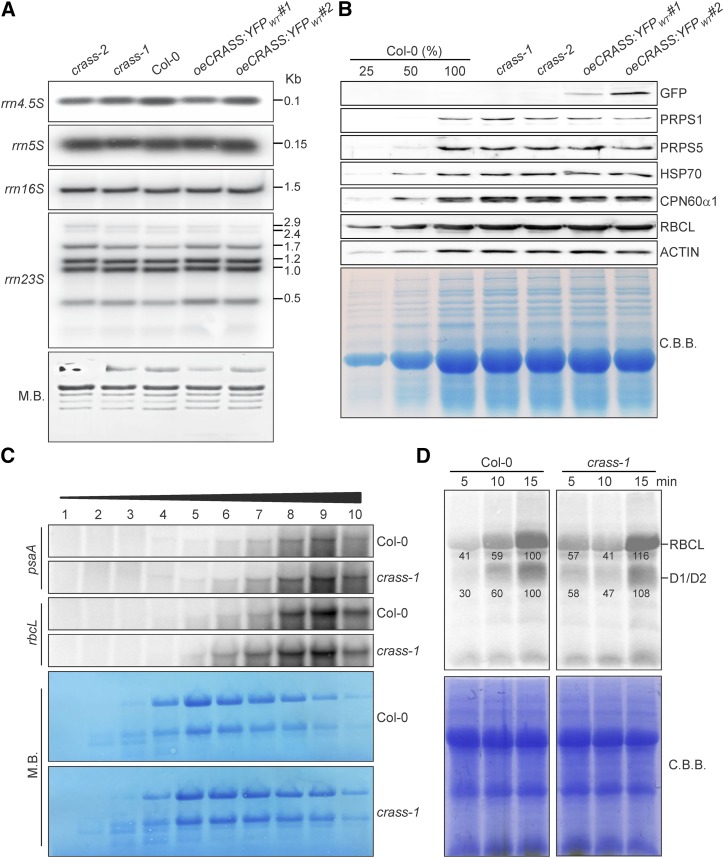
Figure 3.
CRASS is not required for chloroplast translational activity under nonstressed conditions. A, RNA gel-blot analysis of 10-μg samples of total RNA from 3-week-old wild-type (Col-0), mutant (crass-1 and crass-2), and overexpressor (oeCRASS:YFPWT#1 and oeCRASS:YFPWT#2) lines with probes specific for the four plastid rRNAs (rrn23, rrn16, rrn5, and rrn4.5). The sizes of the transcripts are given in kb on the right. M.B., Methylene Blue. B, The steady-state levels of representative chloroplast proteins were analyzed by immunoblotting of samples isolated from 3-week-old plants. Total protein extracts from the same genotypes as in A were examined (loading equal amounts of proteins), together with a dilution series of the Col-0 sample as indicated. Representative images of immunoblots probed with antibodies specific for the indicated proteins are shown. Quantification of the results of five biological replicates (by ImageJ) relative to Col-0 plants is provided in Supplemental Table S1. C, CRASS is not required for efficient polysome loading. RNA gel-blot analysis is shown for psaA and rbcL transcripts in polysome fractions 1 to 10 collected after Suc-gradient centrifugation of wild-type (Col-0) and crass-1 extracts (see “Materials and Methods”). rRNA was stained with Methylene Blue. D, Pulse-labeling analysis of D1/D2 synthesis. Leaves isolated from plants at the six-leaf rosette stage were pulse labeled with [35S]Met under low-light illumination (20 μmol photons m−2 s−1) for 5, 10, and 15 min in the presence of cycloheximide to inhibit cytosolic protein synthesis. Total leaf proteins were then isolated, fractionated by SDS-PAGE, and detected by autoradiography. A portion of the SDS-polyacrylamide gel corresponding to the RBCL region was stained with Coomassie Brilliant Blue (C.B.B.) and served as an internal standard for loading normalization. Quantification of signals (by ImageJ) relative to Col-0 at the 15-min time point (=100%) is provided below each relevant band.