Figure 2. Structural basis of how K187 and L195 substitutions control hcGAS activity.
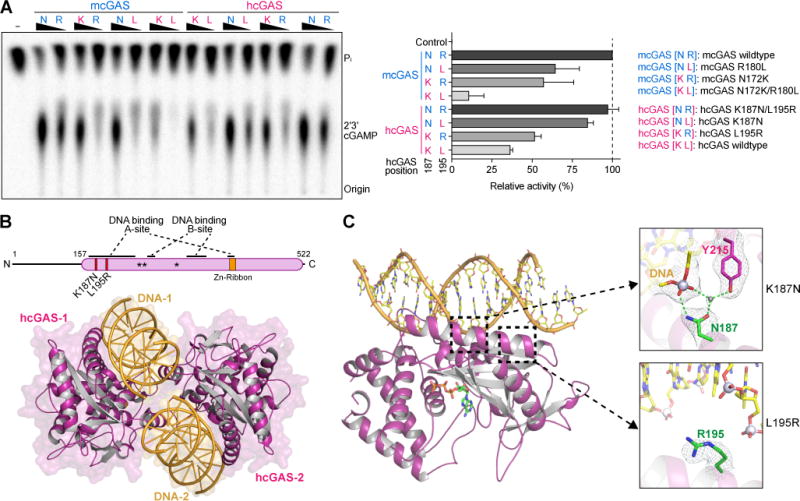
(A) In vitro analysis of the role of hcGAS K187 and L195 variation in 2′3′ cGAMP synthesis regulation. Human and mouse amino acid sequences at 187 (human K187 and mouse-equivalent N187, denoted K or N) and 195 (denoted L or R) were analyzed in both hcGAS and mcGAS backgrounds. Enzymes were stimulated with 45 bp DNA, and 2′3′ cGAMP synthesis was measured as in Figure 1A and quantified relative to maximal activity observed with mcGAS. Data are represented as mean ± SD of three independent experiments.
(B) Schematic and overview of the hcGAS–DNA complex. hcGAS forms a 2:2 complex with DNA where each cGAS monomer has two distinct DNA-binding surfaces (DNA A-Site and DNA B-Site). Stars in the schematic denote the enzyme metal-coordinating active-site residues, schematic not to scale.
(C) Overview of a single 1:1 cGAS–DNA unit in the hcGAS–DNA–ATP complex. Zoom-in cutaways of the locations of K187 and L195 human-specific cGAS substitutions in the DNA A-site. The water molecule coordinated by the K187N mutation and Y215 is depicted as a grey sphere. See also Table S1 and Figure S4.
