Abstract
The cellular uptake of dsRNA after dietary exposure is critical for RNAi efficiency; however, the mechanism of its uptake in many insects remains to be understood. In this study, we evaluated the roles of the endocytic pathway genes Clathrin heavy chain (Chc), Clathrin adaptor protein AP50, ADP ribosylation factor-like 1 (Arf72A), Vacuolar H+ ATPase 16 kDa subunit (Vha16), and small GTPase Rab7 and putative sid-1-like genes (silA and silC) in RNAi response in western corn rootworm (WCR) using a two-stage dsRNA exposure bioassay. Silencing of Chc, Vha16, and AP50 led to a significant decrease in the effects of laccase2 dsRNA reporter, indicating that these genes are involved in RNAi response. However, the knockdown of either Arf72A or Rab7 did not suppress the response to laccase2 dsRNA. The silencing of the silC gene did not lead to a significant reduction in mortality or increase in the expression of V-ATPase A reporter. While the silencing of the silA gene significantly decreased insect mortality, significant changes in V-ATPase A expression were not detected. These results suggest that clathrin-dependent endocytosis is a biological mechanism that plays an important role during RNAi response in WCR adults. The fact that no definitive support for the roles of silA or silC in RNAi response was obtained support the idea that RNAi response varies greatly in different insect species, demanding additional studies focused on elucidating their involvement in this mechanism.
Introduction
Experiments conducted in several insect orders have shown that RNAi can be used as a potential tool for insect pest management by induction of an RNAi response after ingestion of dsRNA [1–7]. The mechanism that allows uptake of the dsRNA from the gut lumen by midgut cells and the subsequent systemic spread from cell to cell is likely to have significant influence on the efficiency of RNAi. Considering that the level of RNAi response across insect orders is highly variable [8–10], a better understanding of this process could aid in the development and improvement of RNAi-based technologies for insect pest management.
The western corn rootworm (WCR), Diabrotica virgifera virgifera LeConte, is one of the most important insect pests of maize throughout the U.S. Corn Belt [11]. Control failures of WCR with synthetic insecticides, crop rotation, and transgenic plants expressing Bacillus thuringiensis toxins have become a serious problem and new management strategies are urgently needed [12–18]. To complement current management strategies, transgenic maize events based on RNAi for WCR control are likely to be deployed by the end of the decade [7].
WCR exhibits a robust systemic RNAi response induced by direct injection of dsRNA [19], feeding of dsRNA provided in artificial diet [20–22], or transgenic plants expressing dsRNA [2, 20]. Understanding the mechanism of dsRNA uptake and the spread of the RNAi signal will contribute not only to the improvement of RNAi efficiency for the management of WCR and other insect pests but will also generate valuable information about potential and some specific mechanisms of insect resistance to RNAi-based technologies.
The systemic RNA interference-defective-1 (SID-1) protein is perhaps the best studied factor required for dsRNA uptake and systemic RNAi [23, 24]. In Caenorhabditis elegans, the SID-1 multi-transmembrane domain protein is present in the cell membrane and functions as a channel for the passive transport of dsRNA between cells [23]. In addition to SID-1, other proteins are involved in dsRNA uptake in C. elegans, including SID-2, SID-3, and SID-5. SID-2 is responsible for the active import of environmental dsRNA from the intestinal lumen [25, 26], while SID-3 is a conserved tyrosine kinase that allows the entry of dsRNA into the cells [27]. In contrast, SID-5 is an endosome-associated protein, believed to be involved in the release of dsRNA from the endosome and movement of the RNAi signal from cell to cell [28, 29].
Homologous sequences of the sid-1 gene have been identified in many species of different insect orders, except for Diptera [30]. In WCR, only two sid-1-like genes have been found and the phylogenetic analysis of the deduced proteins indicates that they are orthologous to the Tribolium silA and silC genes [31]. An association between the presence of sid-1-like genes and the systemic spread of dsRNA was initially proposed since sid-1-like genes are absent in Drosophila melanogaster and other dipterans, species that do not display a robust systemic RNAi response. However, subsequent studies on other insect orders demonstrated that the presence of sid-1-like genes does not necessarily result in a robust systemic RNAi response. For example, the silkworm, Bombyx mori, harbors three sid-1-like genes, but it does not exhibit a substantial systemic RNAi response [30].
Endocytosis has been suggested as an alternative dsRNA uptake mechanism in D. melanogaster S2 cells [32, 33]. The silencing of the Chc gene, which plays a critical role in clathrin-dependent endocytosis, reduced the lethality of D. melanogaster S2 cells after exposure to Ubiquitin (Ubi-p63E) dsRNA, an essential gene for cell viability [33]. A functional screening of a dsRNA library from D. melanogaster S2 cells found several genes involved in the endocytic pathway [32], including Clathrin heavy chain (Chc), Clathrin adaptor protein AP50, that is also known as the μ subunit of the AP2 adapter complex (AP-2μ/AP50) [34], ADP ribosylation factor-like 1 (Arl1/Arf72A) [35], Vacuolar H+ ATPase (V-ATPase) 16 kDa (CG3161) and SFD (CG17332) subunits, and small GTPase Rab7 to be necessary for cellular uptake of dsRNA [32]. The proteins encoded by these genes have been associated with various functions in the clathrin-dependent endocytosis. Chc is the major component of clathrin-coated pits formed on the inner surface of the cytoplasmic membrane [34]; AP50 acts linking the clathrin to its cargo [34] and Arl1/Arf72A is involved in endosomal trafficking at trans-Golgi [36, 37], while Vacuolar H+ ATPase (Vha16) and Rab7 play critical roles in endosome maturation by acidifying endosomes and promoting late endosome fusion, respectively [32, 38].
The requirement of the endocytic pathway for cell entry of dsRNA has also been suggested in other insects including the desert locust, Schistocerca gregaria [39], the oriental fruit fly, Bactrocera dorsalis [40], the red flour beetle, Tribolium castaneum [41], and Colorado potato beetle, Leptinotarsa decemlineata [42, 43]. The silencing of different genes directly related to clathrin-dependent endocytosis, including, Chc, Arf72A, Rab7, AP50, and Vacuolar H+ ATPase SFD subunit (VhaSFD) was able to block significantly the RNAi response in T. castaneum, supporting the involvement of endocytosis in dsRNA uptake [41]. Similarly, the knockdown of Chc and Vacuolar H+ ATPase 16 kDa subunit (Vha16) in S. gregaria also diminished the RNAi response in this insect [39].
While it is possible that both clathrin-dependent endocytosis and the SIL proteins are involved in the RNAi effect and the uptake/spread of the dsRNA in WCR, a direct comparison of their impact on RNAi response will help to identify key genes involved in these processes and the reasons for a potential diverse uptake/spread response during RNAi mechanism. By testing both endocytic genes and sil genes in L. decemlineata [42], it was established that endocytosis is the major contributor to successful RNAi in this insect. Since WCR is a major agricultural pest that is readily susceptible to RNAi and is a robust laboratory model, elucidating key pathways involved in its RNAi response can pave the way to understanding of environmental RNAi in other insects.
In the present study, we report that interruption of the endocytic pathway by silencing AP50, Chc, and Vha16 genes decreased the suppression of the non-lethal reporter gene, laccase2, demonstrating that clathrin-dependent endocytosis is involved in RNAi response in WCR adults. In contrast, the knockdown of sil genes yielded mixed results. silA knockdown generated a reduced RNAi response in WCR adults exposed to dsRNA of the lethal reporter gene, V-ATPase A, that could not be supported at the transcript level. The knockdown of silC or both sil genes simultaneously did not affect the RNAi response. These results do not provide a strong evidence for the involvement of sil genes in the RNAi response in WCR, yet they do not rule it out.
Material and methods
Insects and diet
Newly emerged non-diapausing WCR adults were purchased from Crop Characteristics Inc. (Farmington, MN). The artificial diet used in all bioassays was modified from Branson and Jackson [44]. The diet consisted of 6 grams of the dry ingredients reported by Branson and Jackson [44], 12.5 ml of water, 0.365 g of agar, 0.7 ml of glycerol, and 27.5 μl of a solution of 47% propionic acid and 6% phosphoric acid to reduce microbial contamination. The diet was dispensed into a Petri dish, allowed to solidify at room temperature, and diet plugs were cut using a cork borer (4 mm diameter). WCR adults were kept in a growth chamber at 23 ± 1°C, 75 ± 5% relative humidity with 16:8 photoperiod.
Identification of endocytic pathway genes and putative sid-1-like in the WCR transcriptome
Nucleotide sequences encoding Clathrin heavy chain (Chc) (KJ135005.1) and Vacuolar H+ ATPase 16kDa subunit (Vha16) (KJ135006.1) from S. gregaria were used as query sequences to search for putative homologs in the WCR transcriptome. Putative sid-1-like (silA and silC) and clathrin-mediated endocytosis genes were identified by BLAST searching the WCR transcriptome database described by Eyun et al. [45], using the reported sequences from S. gregaria and T. castaneum [30, 39, 41]. The sequences encoding Clathrin adaptor protein AP50 (AP50) (KJ476827), small GTPase Rab7 (Rab7) (KJ476829), ADP-ribosylation factor-like protein 1 (Arf72A) (XM_967932.3), systemic RNA interference defective-1-related A (Sil-1A) (NM_001105542.1), and systemic RNA interference defective-1-related C (Sil-1C) (NM_001105658.1) from T. castaneum were used as queries. The identified putative WCR orthologs [Chc (KX965603), Vha16 (KX965604), AP50 (KX965605), Arf72A (KX965607), Rab7 (KX965606), silA (KX965608) and silC (KX965609)] were deposited in GenBank.
Double stranded RNA (dsRNA) preparation
Total RNA was isolated from the whole body of a single WCR adult using GeneJET RNA Purification Kit (Fermentas-Thermo Scientific, Waltham, MA) following manufacturer’s instructions. First-strand cDNA was generated with 500 ng of total RNA using the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA). Gene-specific primers were designed with Primer3Plus software [46, 47] to amplify silA, silC, Chc, Vha16, AP50, Arf72A, Rab7, laccase2, and GFP gene. All primers included the T7 promoter sequence at the 5’ end. The primers described by Rangasamy and Siegfried [22] were used for the amplification of V-ATPase A gene (S1 Table). All PCR amplification products were sequenced to confirm the identity and specificity. For negative control, the non-specific green fluorescence protein gene (GFP) was amplified from the pIZT/V5-His expression vector (Invitrogen, USA).
The amplified PCR products were purified using QIAquick PCR Purification Kit (Qiagen, Valencia, CA) and used as templates for in vitro dsRNA synthesis using the MEGAscript high-yield transcription Kit (Applied Biosystems Inc., Foster City, CA). The synthesized dsRNAs were purified using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions, quantified using NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Franklin, MA), and then examined by agarose gel electrophoresis to determine their purity and integrity.
Insect bioassay for functional analysis of endocytic genes
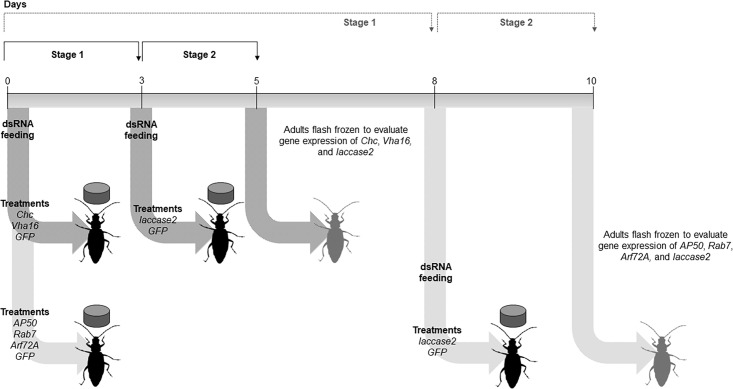
To determine the involvement of the clathrin-dependent endocytosis in dsRNA uptake in WCR, we used a similar approach to that described by Velez et al. [48]. However, we modified the time between the first and second dsRNA exposure to account for time necessary to generate gene silencing and the effect of the gene silencing on the survival of WCR adults. The bioassays with Chc and Vha16 were performed in five days since silencing of these genes affected the survival of insects, with the first and second dsRNA exposure on day zero and day three.
The bioassays with AP50, Arf72A, and Rab7 were performed over ten days since these genes had weak knockdown after three days of exposure to dsRNA. The first and second dsRNA exposures occurred on days zero and eight (Fig 1). In this experiment, both dsRNA exposures were performed by feeding. During the first dsRNA exposure, WCR adults were fed with diet plugs treated with 600 ng of Chc, Vha16, AP50, Arf72A, Rab7, or GFP dsRNA every other day. For the second dsRNA exposure, new diet plugs coated with 600 ng of laccase2 or GFP dsRNA were provided to WCR adults on day three or eight, depending on the group treatment. One WCR adult was collected per replication from all the treatments immediately before and two days after the second dsRNA exposure, flash-frozen in liquid nitrogen, and stored at -80°C for total RNA extraction. Each assay was performed in triplicate with a total of 14 beetles per treatment.
Fig 1. Two-step bioassay to determine the effect of the suppression of the endocytic genes on the expression of laccase2 in WCR adults.
Insect bioassay for functional analysis of silA and silC genes
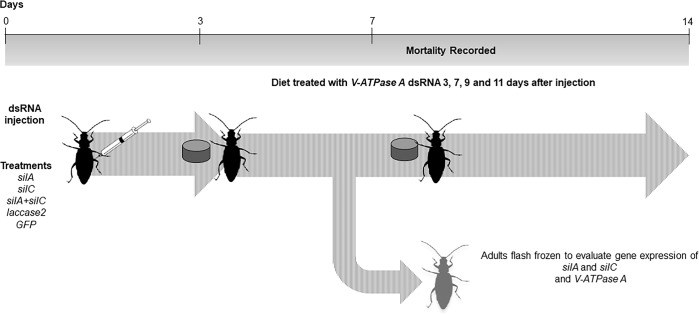
To determine if the lethal effect of the V-ATPase A dsRNA would be altered by the knockdown of sil genes we used a similar approach to that described by Velez et al. [48] with slight modifications. Briefly, a volume of 0.6 μl of silA and silC dsRNA at 1 μg/μl, and a mixture of silA and silC dsRNA containing equal amounts of both dsRNAs, for a total of 600 ng, were injected into individual beetles. Insects were injected between the coxae of the last pair of legs using a glass capillary syringe and then fed with untreated diet.
To determine if competitive inhibition of dsRNA occurred when multiple species-specific dsRNAs were provided to beetles, WCR adults were also injected with dsRNA targeting the non-lethal gene laccase2, which is required for cuticular pigmentation [49]. WCR adults injected with GFP dsRNA were used as controls and un-injected adults were used to evaluate the mortality associated with the injury caused by the injection. The beetles were anesthetized with carbon dioxide to facilitate injections.
For the secondary exposure to dsRNA, diet plugs (4 mm diameter x 2 mm height) were surface-treated with 500 ng of V-ATPase A dsRNA and provided to beetles three, five, seven, nine, and eleven days after injection. For the remainder of the assays, beetles were provided with untreated diet for a total of 14 days (Fig 2). On day seven, one beetle per replication was collected from all the treatments, flash frozen in liquid nitrogen and stored at -80°C for total RNA extraction. A total of 16 beetles were used in each treatment and mortality was recorded daily until day 14. Each assay was performed in triplicate.
Fig 2. Relative transcript levels of the endocytosis-related genes after dsRNA exposure.
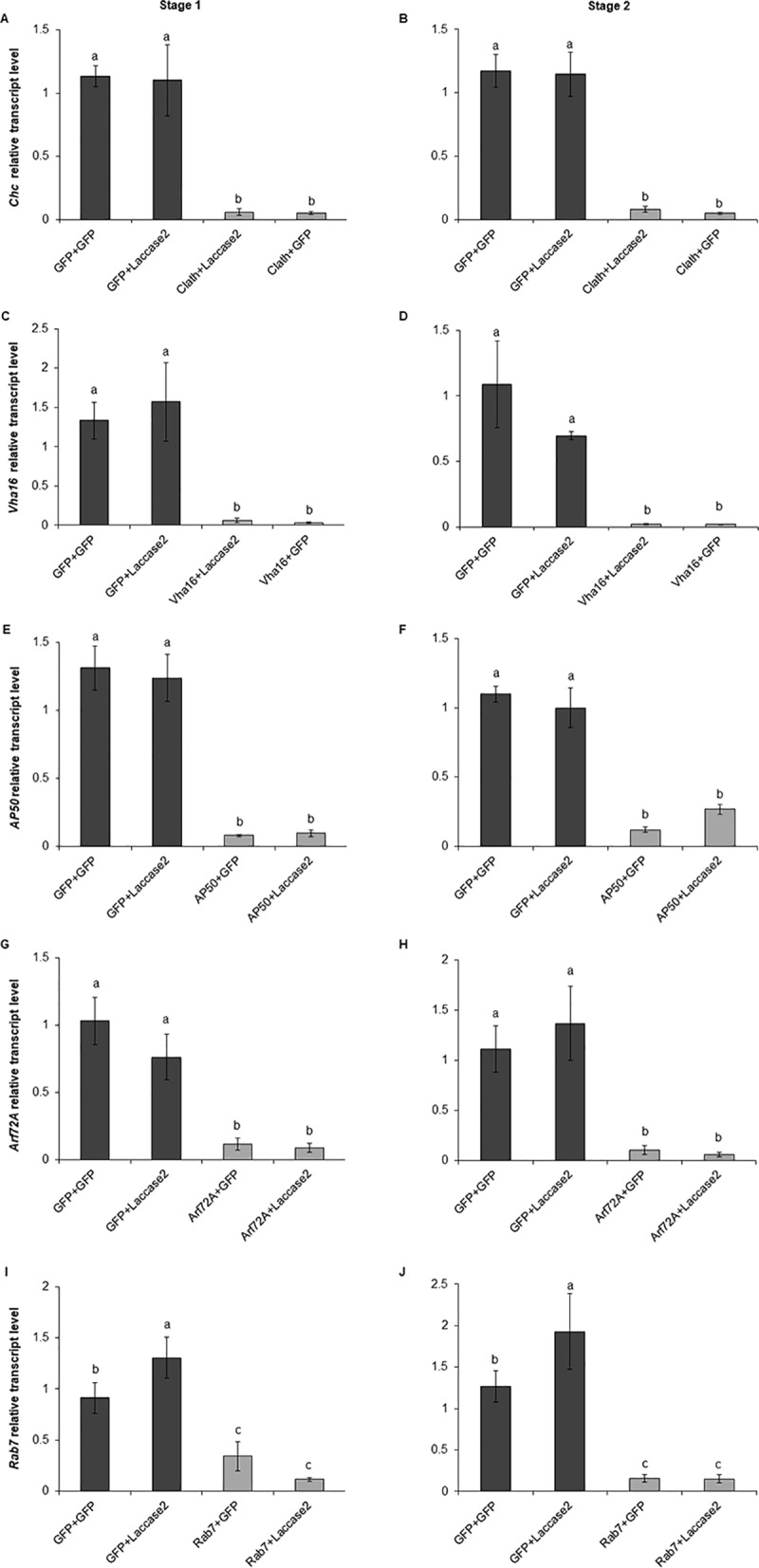
Relative transcript levels of Chc (A and B), Vha16 (C and D), AP50 (E and F), Arf72A (G and H), and Rab7 (I and J) genes in WCR adults after the first (stage 1) and second (stage 2) dsRNA exposure evaluated by RT-qPCR. Values shown are the means and standard errors (±SE) of three biological replicates each with two technical replicates. Different letters represent significant differences at p-value < 0.05.
Quantitative Real Time PCR (RT-qPCR)
Total RNA was isolated from the whole bodies of adults using GeneJET RNA Purification Kit (Fermentas-Thermo Scientific, Waltham, MA) following the manufacturer’s recommendations. cDNA was synthetized with the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA) using 500 ng of RNA, following the manufacturer’s instructions. The RT-qPCR reactions included 1 μl of cDNA diluted 50X, 5 μl of Fast SYBR® Green Master Mix (Applied Biosystems, Foster City, CA), 0.2 μl at 10 μM of each primer, and 3.6 μl of nuclease-free water, for a total volume of 10 μl. The primers were designed with Primer3Plus [46, 47] and validated by PCR amplification efficiencies (E) and correlation coefficients (R2) analysis (S2 Table).
Both primer efficiency test and RT-qPCR were performed on a 7500 Fast RT-PCR System (Applied Biosystems, Grand Island, NY). The thermocycler conditions were one cycle at 95°C for 20 s, followed by 40 cycles of denaturation at 95°C for 3 s and annealing/extension at 60°C for 30 s. At the end of each PCR reaction, a melting curve was generated to confirm a single peak and rule out the possibility of primer-dimers and nonspecific product formation. The expression of the genes was calculated using the 2-ΔΔCT method [50]; using actin as reference gene which has previously demonstrated to be stable under experimental conditions in which WCR were exposed to dsRNA treatments [51]. RT-qPCR analysis was performed with three biological replicates and two technical replicates.
Statistical analysis
Gene expression and mortality were subjected to an analysis of variance (ANOVA) using the PROC GLIMMIX procedure; least-squares differences were used to perform pairwise comparisons between treatments. All statistical analyses were performed using SAS Software version 9.4 [52]. Data were expressed as mean ± standard error of the mean (SE), and values of p < 0.05 were considered statistically significant.
Results
Identification of putative clathrin-dependent endocytic pathway and sid-1-like genes in the WCR transcriptome
A BLAST search allowed the identification of putative Chc (NCBI ID: KX965603), Vha16 (KX965604), AP50 (KX965605), Arf72A (KX965607), Rab7 (KX965606), silA (KX965608) and silC (KX965609) genes in the WCR transcriptome database. We also searched for the putative silB gene, however, no orthologs to the Tribolium silB gene were found in the WCR transcriptome. Additional BLASTX analysis revealed that the deduced WCR protein sequences display high sequence identity to their orthologs from different insect species and shared the highest identity with proteins from T. castaneum or L. decemlineata [above 67%] (S3 Table). A reverse BLAST of the C. elegans proteome (http://www.wormbase.org/) using the putative WCR SILA and SILC proteins has revealed CHolesterol UPtake associated protein CHUP-1 as the closest homolog in C. elegans (S1 Fig).
Knockdown of clathrin-dependent endocytosis elements impedes WCR’s RNAi response
To examine the hypothesis that the clathrin-dependent endocytosis is involved in RNAi response in WCR adults, we used an “RNAi-of-RNAi” approach (Fig 1), which has been successfully used to associate Dicer-2 and Argonaute 2 (AGO2) with the RNAi pathway in WCR [48]. For this purpose, genes encoding proteins active in steps of the endocytic pathway and possibly involved in RNAi response in other insects [32, 33, 39–43] were silenced, and the effect on laccase2 gene knockdown was analyzed.
Gene knockdown of Chc, Vha16, AP50, Arf72A, and Rab7 was evaluated in WCR adults after the first dsRNA exposure (stage 1) and two days after the second dsRNA exposure (stage 2) (Fig 1). In this experiment, lacasse2 was used as the reporter gene instead of V-ATPase A as reported by Velez et al. [48], since Chc and Vha16 dsRNA generate mortality, which could lead to misinterpretation of RNAi-derived phenotype. Moreover, both the V-ATPase A subunit (an ortholog of Vha68-2) and Vha16 encode subunits of the same functional protein complex [53], hence V-ATPase A was not used as a reporter. The expression levels of all five target genes were strongly reduced after the first dsRNA exposure and remained suppressed after the second dsRNA exposure to laccase2 or GFP dsRNA (Fig 2). These results suggest that the expression of the endocytic genes was unaffected by the exposure to a second dsRNA and the knockdown did not decrease during the time that the WCR adults were exposed to the second dsRNA.
In WCR adults fed with GFP dsRNA and later with laccase2 dsRNA, the lacasse2 expression decreased by 79.4%, relative to the control in which WCR adults were treated at both stages with GFP dsRNA. In contrast, when WCR adults were exposed to AP50 dsRNA, followed by laccase2 dsRNA, the laccase2 expression was reduced by only 5.9% (Fig 3A). This result strongly suggests that the down-regulation of AP50 significantly antagonizes the knockdown of laccase2, indicating that AP50 plays an important role in the RNAi response in WCR adults.
Fig 3. Effect of the knockdown of the endocytosis-related genes on the RNAi response.
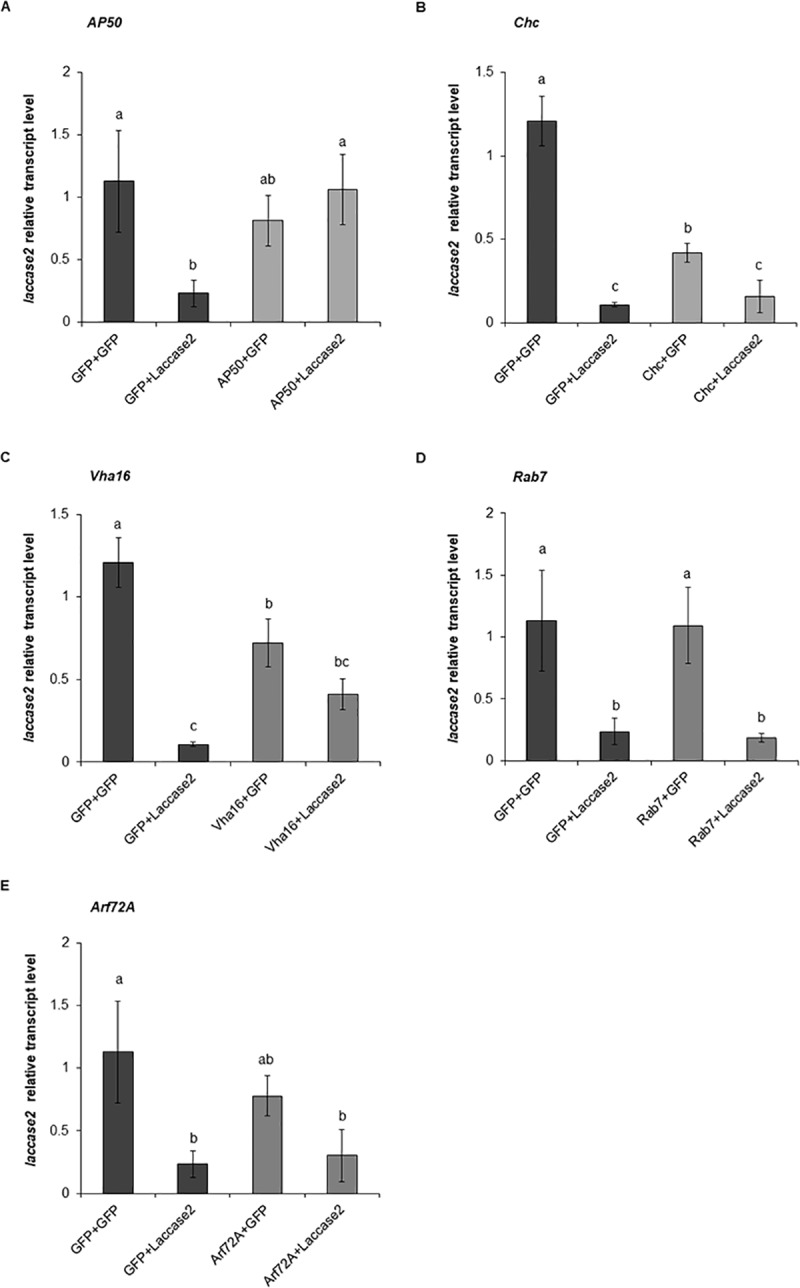
Effect of the knockdown of the AP50 (A), Chc (B), Vha16 (C), Rab7 (D), and Arf72A (E) on the relative transcript level of laccase2 was evaluated by RT-qPCR analysis. Values shown are the means and standard errors (±SE) of three biological replicates each with two technical replicates. Different letters represent significant differences at p-value < 0.05.
We did not observe significant increase in laccase2 expression when the WCR adults were fed with Chc or Vha16 dsRNAs and subsequently treated with laccase2 dsRNA compared to beetles fed with GFP dsRNA and later with laccase2 dsRNA (Fig 3B and 3C). However, it was observed that both Chc and Vha16 dsRNAs caused a reduction in laccase2 transcript levels, 65.3% and 40.4%, respectively (Fig 3B and 3C). Therefore, it appears that the knockdown of Chc and Vha16 genes also affected the expression of the reporter gene. Similar effects were observed in Metaseiulus occidentalis and Locusta migratoria, in which Chc silencing reduced the expression of the reporter genes, cathepsin and methoprene-tolerant by 40% and 88%, respectively [54].
Consequently, to prevent confounding results from the Chc+laccase2 and Vha16+laccase2 treatments by this non-specific effect, we subsequently used a different approach, as described by Wu and Hoy [54], for the calculation of the reporter gene knockdown in order to evaluate the effect of Chc and Vha16 silencing in laccase2 expression. In this approach, Chc+GFP and Vha16+GFP were used as controls to calculate laccase2 knockdown in Chc+laccase2 and Vha16+laccase2 treatments.
The gene silencing of laccase2 in insects that were first treated with GFP dsRNA, followed by laccase2 dsRNA (GFP+GFP and GFP+laccase2) was 91.1% (Fig 3B). Insects treated with Chc dsRNA and subsequently with laccase2 dsRNA (Chc+GFP x Chc+laccase2) showed a reduction of laccase2 expression by 62.5% (Fig 3B), while in beetles treated with Vha16 dsRNA and later with laccase2 dsRNA (Vha16+GFP and Vha16+laccase2) the laccase2 expression decreased by 43.0% (Fig 3C). These data indicate that Chc and Vha16 transcript suppression reduced the expression of laccase2 by 28.6% and 48.1%, respectively (Fig 3B and 3C); reinforcing our hypothesis that clathrin-dependent endocytosis is required for RNAi response in WCR adults. The Arf72A and Rab7 silencing, however, did not result in significant retention of laccase2 expression (Fig 3D and 3E).
The effect of silA and silC genes silencing on RNAi response
To determine if the SIL proteins are involved in the RNAi response in WCR adults, we performed experiments using an “RNAi-of-RNAi” approach described by Velez et al. [48] (Fig 4). In a previous study, feeding of WCR adults on artificial diet treated with dsRNA of the Vacuolar-ATPase subunit A (V-ATPase A) gene caused high mortality after 14 days of exposure [22]. Therefore, we chose V-ATPase A as a reporter gene to investigate the role of the SILA and SILC proteins in the RNAi response in WCR adults (Fig 4). The WCR V-ATPase A is most closely related to Drosophila melanogaster Vha68-2 (CG3762), and the predicted V-type proton ATPase catalytic subunit A (XP_976188.1) of Tribolium castaneum. To determine if competition for the RNAi machinery occurred between the dsRNAs from the first and the second exposures, a treatment group in which the WCR adults were first injected with laccase2 dsRNA, followed by feeding with V-ATPase A dsRNA was included. Lacasse2 is involved in cuticular tanning [49] and is not associated with RNAi response.
Fig 4. Two-step bioassay to determine the effect of the suppression of silA and silC genes on the mortality and V-ATPase A expression in WCR adults.
Seven days after dsRNA injections a robust reduction in expression of the silA, silC, and laccase2 genes was observed (Fig 5). Knockdown of silA and silC genes was achieved by injection of dsRNA rather than feeding, as we have previously observed higher and more consistent rates of knockdown using an injection. In the treatment group silA+V-ATPase A and silA/silC+V-ATPase A, the expression of silA was significantly reduced compared to the control treatment (adults injected with GFP dsRNA and subsequently fed with V-ATPase A dsRNA) by 89.1% and 87.5%, respectively (Fig 5A). The expression of silC in the treatment group silC+V-ATPase A and silA/silC+V-ATPase A was reduced by 83.9% and 95.5%, respectively, and was significantly different from the control (Fig 5B). Additionally, the reduction of laccase2 expression was 98.8% and 99.6% when the WCR adults were injected with laccase2 dsRNA and subsequently fed with untreated diet or V-ATPase A dsRNA, respectively (Fig 5C).
Fig 5. Relative transcript level of silA, silC and laccase2 genes after dsRNA exposure.
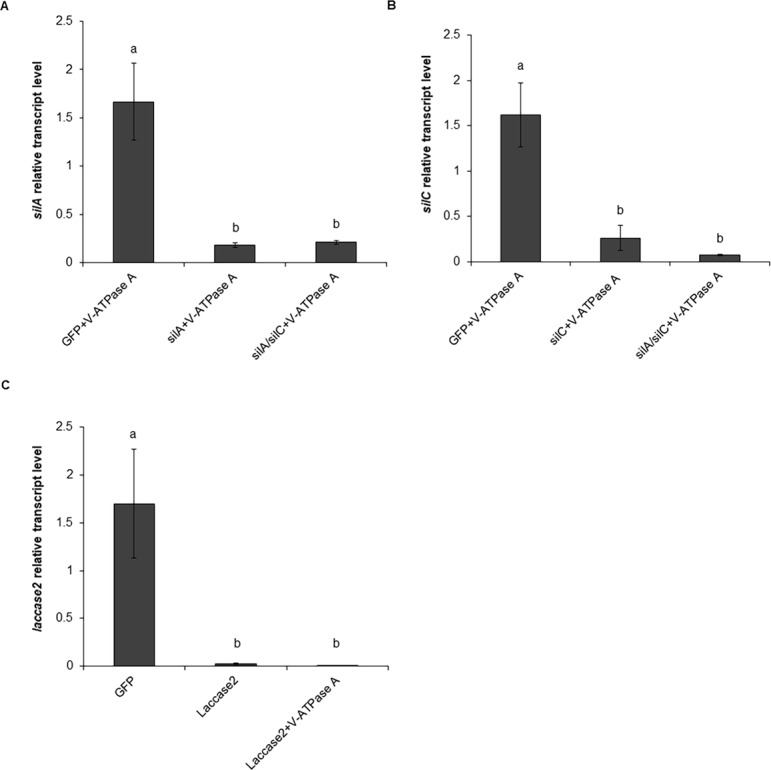
Relative transcript level of silA (A), silC (B) and laccase2 (C) evaluated seven days after the injection of 600 ng of the respective dsRNA or a combination of silA and silC dsRNAs into WCR adults. Values shown are the means and standard errors (±SE) of three biological replicates, each with two technical replicates. Different letters represent significant differences at p-value < 0.05.
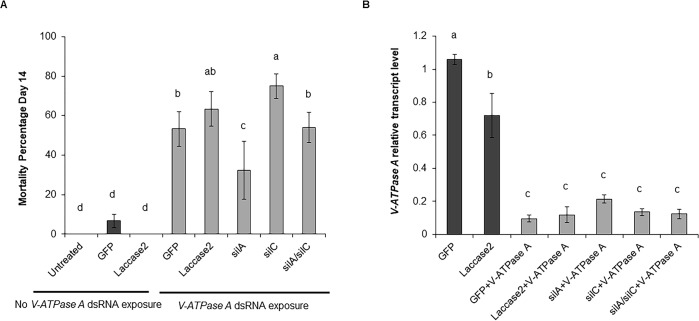
The knockdown of laccase2 did not affect the knockdown of the V-ATPase A gene or the mortality of WCR adults, when compared with control insects injected with GFP dsRNA and fed with V-ATPase A dsRNA (Fig 6A and 6B). These results indicate that mortality and V-ATPase A expression of the treatment groups injected with sil dsRNA and fed with V-ATPase A dsRNA were not due to potential competition between dsRNAs. Furthermore, the concurrent silencing of silA and silC genes suggests that oversaturation of the RNAi machinery did not occur when a mixture of silA and silC dsRNAs was injected, at least at dsRNA concentrations that were evaluated in this assay (Fig 5A and 5B).
Fig 6. Mortality of WCR adults and relative V-ATPase A expression.
(A) Mortality of WCR adults from the different treatment groups after 14 days. WCR adults were injected with 600 ng of GFP, laccase2, silA, silC, or a combination of silA and silC dsRNAs and subsequently fed with V-ATPase A dsRNA. WCR adults injected with GFP and not exposed to V-ATPase A dsRNA were used as controls. (B) Relative V-ATPase A transcript levels were evaluated by RT-qPCR seven days after exposed to V-ATPase A dsRNA. Values shown are the means and standard errors (±SE) of three biological replicates. Different letters represent significant differences at p-value < 0.05.
The down-regulation of the silC gene did not significantly suppress the mortality of the WCR adults, which were injected with silC dsRNA or with the mixture of silA and silC dsRNAs and subsequently fed with V-ATPase A dsRNA, compared to the control (Fig 6A). Furthermore, V-ATPase A transcript abundance was not significantly influenced by the down-regulation of the silC, implying that in WCR adults, SILC protein is not required for effective RNAi response (Fig 6B). When WCR adults were injected with silA dsRNA and fed with V-ATPase A dsRNA, no significant increase in V-ATPase A transcript abundance was observed. However, the mortality of the WCR adults was significantly reduced (Fig 6A), suggesting that the SILA protein may play a role in the RNAi response.
Discussion
Most studies aimed at understanding the mechanisms of cellular dsRNA uptake and the systemic spread of RNAi effect in insects have focused on SIL proteins and the endocytic pathway as key participants [31–33, 39–43]. Since the level of environmental RNAi response varies greatly from one insect to another, the genes involved in this response may also vary. To elucidate which gene products are involved in dsRNA uptake in WCR, the present work investigated the impact of knocking down several components of clathrin-dependent endocytosis and homologs of SID-1 on the RNAi response in WCR beetles. The genes evaluated in this study included WCR endocytosis-related targets Chc, Vha16, AP50, Arf72A, Rab7 and SID-like genes SilA and SilC.
Role of the endocytic pathway in dsRNA uptake has been demonstrated in D. melanogaster S2 cells, and confirmed in S. gregaria, L. decemlineata, T. castaneum and B. dorsalis, suggesting that this mechanism of dsRNA internalization might be widespread among insects [32, 33, 39–42]. A study by Ulvila et al. [33] confirmed that Chc was necessary for dsRNA uptake in Drosophila S2 cells, by bypassing this pathway via transfection. Further, studies in S2 cells [32] and T. castaneum [41] have confirmed the involvement of clathrin-mediated endocytosis in RNAi response by pharmacological block via bafilomycin-A1 and bafilomycin-A1 or chlorpromazine, respectively. Xiao et al. [41] showed that both bafilomycin-A1 and chlorpromazine can block the uptake of fluorescently-labeled dsRNA by larval midgut cells. While by their nature, the RNAi-of-RNAi experiments performed in the current study cannot identify which part of RNAi process is perturbed, by extension, the genes identified in our study as key participants in RNAi response are likely to be involved in dsRNA uptake.
In the present study, knockdown of WCR endocytic genes, AP50, Chc, and Vha16 reduced the subsequent knockdown of the reporter gene laccase2 (Fig 3A–3C), confirming that the endocytic pathway participates in the RNAi response. Results from this study support the observations in coleopteran insects L. decemlineata and T. castaneum, where silencing of endocytosis-related genes Chc and Vha16, reduced the subsequent RNAi response [41–43]. Anchoring on AP50, Chc, and Vha16 results, we identified clathrin-mediated endocytosis as being essential for a robust RNAi response in WCR.
In contrast to the robust effects of AP50, Chc, and Vha16 on RNAi response in WCR, knockdown of Arf72A and Rab7 did not result in significant increase of laccase2 expression (Fig 3D and 3E). Although we observed no effect for Rab7 and Arf72A, partial block of the RNAi response was observed in L. decemlineata (Lepd-SL1) cells and T. castaneum when these genes were silenced [41, 43]. Unlike Chc or Vha16, which may be necessary throughout the endocytic cycle, Rab7 is associated with late endosomes/multivesicular bodies. Arf72A/Arl1 is known to associate primarily with the Golgi network [35, 55] and binds AP1 clathrin adaptor [37] (Fig 7). To become available to the RNAi machinery in the cytosol, the dsRNA needs to escape from the early to late endosomes before they fuse with lysosomal compartments [56]. It is hence possible that sufficient laccase2 dsRNA escaped in the earlier steps of the endocytosis, thus the knockdown of Arf72A and Rab7 genes did not significantly affect the expression of laccase2. These hypotheses are further supported by the fact that in coleopteran insects dsRNA escape from the endosomal compartments appears to be an efficient process leading to a robust RNAi response [57].
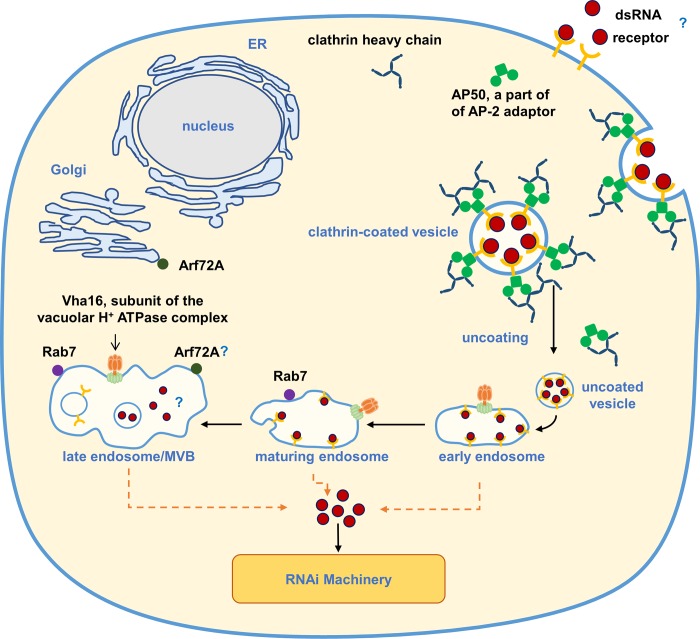
Fig 7. Known components of clathrin-mediated endocytosis involved in the RNAi response in WCR.
The RNAi-of-RNAi approach used in this study implicated Clathrin heavy chain (Chc), Clathrin adaptor protein AP50 (AP50), and Vacuolar H+ ATPase 16 kDa subunit (Vha16) in the RNAi response of WCR. Rab7 is necessary for clathrin-mediated endocytosis, it associates with late endosomes/multivesicular bodies and may function downstream of the dsRNA release into cytoplasm. Arl1/Arf72A associates with trans-Golgi.
In the case of Arf72A, it is possible that its effect on the uptake of dsRNA is indirect, since blocking Arl1/Arf72A causes the dispersion of AP1, which affects secretory granule biogenesis and clathrin exchange [37]. Similarly, depletion of Arl1 protein (Arl1p) in Saccharomyces cerevisiae leads to both decreased protein uptake and secretion since the regulation of membrane traffic is disturbed [58]. Therefore, the block of Arf72A seems to perturb important processes involved in the clathrin pathway, which could interfere in the dsRNA uptake or transport.
The insect studies of SID-like genes have been driven largely by the knowledge from systemic RNAi deficiencies (SID) identified in C. elegans mutagenesis screens. The transmembrane protein systemic RNA interference deficiency-1 (SID-1) is necessary for the uptake of dsRNA in C. elegans, enabling systemic spread of the RNAi effect [24]. The presence of C. elegans sid-1 homologues has been confirmed in multiple insects including Schistocerca americana, Spodoptera exigua, Spodoptera litura, Spodoptera frugiperda, Aphis glycines, Aphis gossypii, Anthonomus grandis, Nilaparva lugens, Apis mellifera, L. decemlineata, and D. v. virgifera larvae [31, 42, 59–67]. However, as observed in our study, the presence of sil genes in insects does not assure the participation of SIL proteins in RNAi response. In our study, only silA significantly suppressed the V-ATPase A knockdown phenotype in WCR adults (Fig 6A). However, this data was not reflected by a commensurate reduction in V-ATPase A transcript levels (Fig 6B). We did not observe an impact of silC on the RNAi response at either transcript or phenotypic levels. Nevertheless, since the nature of RNAi-of-RNAi experiments does not assure full knockdown of the gene targets, it is possible that some SILA or SILC protein remained. Additionally, the half-life of the SIL proteins may allow them to remain functional while their transcripts are largely depleted. Therefore, the participation of D. v. virgifera SILA or SILC proteins in dsRNA response cannot be ruled out.
The participation of the sil genes in the RNAi response has been suggested in N. lugens, L. decemlineata and D. v. virgifera larvae [31, 42, 66]. In L. decemlineata, it has been reported that silA and silC are necessary for an effective RNAi response [42, 43]. Further, the silencing of silA promoted stronger suppression of reporter gene knockdown compared to the silencing of silC [42]. A study performed with WCR larvae demonstrated through an “RNAi-of-RNAi” approach that the silencing of silA and silC genes suppressed the RNAi phenotype of the ebony gene, suggesting the involvement of both sil genes in dsRNA response [31]. However, the phenotype resulting from ebony RNAi was not pronounced [31], suggesting that these proteins probably are not the unique mechanism associated with the uptake of dsRNA in WCR.
Reports for other insects including L. migratoria, S. gregaria, Plutella xylostella and T. castaneum indicate that the sil genes are not involved in systemic RNAi [30, 39, 68, 69]. Detailed sequence analysis of the SIL proteins has revealed that in some insects the N-terminal extracellular domain shares more identity with the TAG-130/CHUP-1 protein of C. elegans, involved with cholesterol internalization, than with the SID-1 protein [30, 68]. We also noted that both WCR SILA and SILC are more similar to the C. elegans cholesterol uptake protein CHUP-1(S1 Fig). Thus, it is possible that the WCR SILA and SILC proteins perform functions associated with cholesterol uptake rather than dsRNA transport [70], and any CHUP-1-associated phenotypes may occur indirectly, through interference with cholesterol endocytic pathway [71] with subsequent impact on membrane transport.
In summary, the comparison of clathrin-mediated endocytosis and sil genes in the RNAi response of WCR suggests that clathrin-dependent endocytosis is more likely to be the primary mechanism for the import of dsRNA in this insect. Further experiments using either WCR biological stages or a coleopteran cell culture similar to those performed by Yoon et al. [43] and Yoon et al. [72] focused on monitoring dsRNA uptake by cells and its movement will help shed light on dsRNA uptake and spread in WCR. Overall, our research provides a starting point for future studies, which surely will have important implications for the development, efficacy, and improvement of RNAi-based management strategies directed to control WCR populations.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was partially supported by CNPq-Conselho Nacional de Desenvolvimento Científico e Tecnológico for DHP’s scholarship (DHP). AMV, HW, AVJ and BDS were funded by the University of Nebraska-Lincoln. Corteva Agriscience™, Agriculture Division of DowDuPont™ provided support in the form of salaries for authors KEN and EF but did not have a role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Bally J, McIntyre GJ, Doran RL, Lee K, Perez A, Jung H, et al. In-plant protection against Helicoverpa armigera by production of long hpRNA in chloroplasts. Frontiers in plant science. 2016;7:1453 10.3389/fpls.2016.01453 PubMed Central PMCID: PMC5040858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, et al. Control of coleopteran insect pests through RNA interference. Nature biotechnology. 2007;25(11):1322–6. 10.1038/nbt1359 [DOI] [PubMed] [Google Scholar]
- 3.Jin S, Singh ND, Li L, Zhang X, Daniell H. Engineered chloroplast dsRNA silences cytochrome p450 monooxygenase, V-ATPase and chitin synthase genes in the insect gut and disrupts Helicoverpa armigera larval development and pupation. Plant biotechnology journal. 2015;13(3):435–46. Epub 2015/03/19. 10.1111/pbi.12355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nature biotechnology. 2007;25(11):1307–13. 10.1038/nbt1352 [DOI] [PubMed] [Google Scholar]
- 5.Ni M, Ma W, Wang X, Gao M, Dai Y, Wei X, et al. Next-generation transgenic cotton: pyramiding RNAi and Bt counters insect resistance. Plant biotechnology journal. 2017;15(9):1204–13. 10.1111/pbi.12709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitino M, Coleman AD, Maffei ME, Ridout CJ, Hogenhout SA. Silencing of aphid genes by dsRNA feeding from plants. PloS one. 2011;6(10):e25709 10.1371/journal.pone.0025709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Khan SA, Hasse C, Ruf S, Heckel DG, Bock R. Pest control. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science. 2015;347(6225):991–4. 10.1126/science.1261680 [DOI] [PubMed] [Google Scholar]
- 8.Baum JA, Roberts JK. Progress towards RNAi-mediated insect pest management. Advances in Insect Physiology. 2014;47:249–95. [Google Scholar]
- 9.Joga MR, Zotti MJ, Smagghe G, Christiaens O. RNAi Efficiency, Systemic Properties, and Novel Delivery Methods for Pest Insect Control: What We Know So Far. Frontiers in physiology. 2016;7:553 Epub 2016/12/03. 10.3389/fphys.2016.00553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, Kanginakudru S, et al. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. Journal of insect physiology. 2011;57(2):231–45. Epub 2010/11/17. 10.1016/j.jinsphys.2010.11.006 [DOI] [PubMed] [Google Scholar]
- 11.Sappington TW, Siegfried BD, Guillemaud T. Coordinated Diabrotica Genetics Research: Accelerating Progress on an Urgent Insect Pest Problem2006 2006-04-01 00:00:00. 90–7 p.
- 12.Devos Y, Meihls LN, Kiss J, Hibbard BE. Resistance evolution to the first generation of genetically modified Diabrotica-active Bt-maize events by western corn rootworm: management and monitoring considerations. Transgenic research. 2013;22(2):269–99. 10.1007/s11248-012-9657-4 [DOI] [PubMed] [Google Scholar]
- 13.Gassmann AJ, Petzold-Maxwell JL, Clifton EH, Dunbar MW, Hoffmann AM, Ingber DA, et al. Field-evolved resistance by western corn rootworm to multiple Bacillus thuringiensis toxins in transgenic maize. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(14):5141–6. 10.1073/pnas.1317179111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine E, Spencer JL, Isard SA, Onstad DW, Gray ME. Adaptation of the western corn rootworm to crop rotation: evolution of a new strain in response to a cultural management practice. American Entomologist. 2002;48:94–107. [Google Scholar]
- 15.Meinke LJ, Siegfried BD, Wright RJ, Chandler LD. Adult susceptibility of Nebraska western corn rootworm (Coleoptera: Chrysomelidae) populations to selected insecticides. Journal of Economic Entomology. 1998;91(3):594–600. 10.1093/jee/91.3.594 [DOI] [Google Scholar]
- 16.Metcalf RL. Implications and prognosis of resistance to insecticides In: Georghiou GP, Saito T, editors. Pest Resistance to Pesticides. New York: Plenum Press; 1983. p. 769–92. [Google Scholar]
- 17.van Rozen K, Ester A. Chemical control of Diabrotica virgifera virgifera LeConte. Journal of Applied Entomology. 2010;134(5):376–84. 10.1111/j.1439-0418.2009.01504.x [DOI] [Google Scholar]
- 18.Wangila DS, Gassmann AJ, Petzold-Maxwell JL, French BW, Meinke LJ. Susceptibility of Nebraska Western Corn Rootworm (Coleoptera: Chrysomelidae) Populations to Bt Corn Events. Journal of Aconomic Entomology. 2015;108(2):742–51. Epub 2015/10/16. 10.1093/jee/tou063 [DOI] [PubMed] [Google Scholar]
- 19.Alves AP, Lorenzen MD, Beeman RW, Foster JE, Siegfried BD. RNA interference as a method for target-site screening in the Western corn rootworm, Diabrotica virgifera virgifera. Journal of Insect Science. 2010;10:162 Epub 2010/11/12. 10.1673/031.010.14122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu X, Richtman NM, Zhao J-Z, Duncan KE, Niu X, Procyk LA, et al. Discovery of midgut genes for the RNA interference control of corn rootworm. Scientific Reports. 2016;6:30542 10.1038/srep30542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Khajuria C, Rangasamy M, Gandra P, Fitter M, Geng C, et al. Long dsRNA but not siRNA initiates RNAi in western corn rootworm larvae and adults. Journal of Applied Entomology. 2015;139(6):432–45. [Google Scholar]
- 22.Rangasamy M, Siegfried BD. Validation of RNA interference in western corn rootworm Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae) adults. Pest Management Science. 2012;68(4):587–91. [DOI] [PubMed] [Google Scholar]
- 23.Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301(5639):1545–7. 10.1126/science.1087117 [DOI] [PubMed] [Google Scholar]
- 24.Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295(5564):2456–9. 10.1126/science.1068836 [DOI] [PubMed] [Google Scholar]
- 25.McEwan DL, Weisman AS, Hunter CP. Uptake of extracellular double-stranded RNA by SID-2. Molecular cell. 2012;47(5):746–54. 10.1016/j.molcel.2012.07.014 PubMed Central PMCID: PMC3488460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winston WM, Sutherlin M, Wright AJ, Feinberg EH, Hunter CP. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(25):10565–70. 10.1073/pnas.0611282104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jose AM, Kim YA, Leal-Ekman S, Hunter CP. Conserved tyrosine kinase promotes the import of silencing RNA into Caenorhabditis elegans cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(36):14520–5. 10.1073/pnas.1201153109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinas A, Wright AJ, Hunter CP. SID-5 Is an Endosome-Associated Protein Required for Efficient Systemic RNAi in C. elegans. Curr Biol. 2012;22(20):1938–43. 10.1016/j.cub.2012.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rocheleau CE. RNA Interference: Systemic RNAi SIDes with Endosomes. Curr Biol. 2012;22(20):R873–R5. 10.1016/j.cub.2012.08.039 [DOI] [PubMed] [Google Scholar]
- 30.Tomoyasu Y, Miller SC, Tomita S, Schoppmeier M, Grossmann D, Bucher G. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008;9(1):R10 10.1186/gb-2008-9-1-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyata K, Ramaseshadri P, Zhang Y, Segers G, Bolognesi R, Tomoyasu Y. Establishing an in vivo assay system to identify components involved in environmental RNA interference in the western corn rootworm. PloS one. 2014;9(7):e101661 Epub 2014/07/09. 10.1371/journal.pone.0101661 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O'Farrell PH, et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nature cell biology. 2006;8(8):793–802. Epub 2006/07/25. 10.1038/ncbl439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, Kocks C, et al. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. The Journal of biological chemistry. 2006;281(20):14370–5. Epub 2006/03/15. 10.1074/jbc.M513868200 . [DOI] [PubMed] [Google Scholar]
- 34.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Bio. 2011;12(8):517–33. 10.1038/nrm3151 [DOI] [PubMed] [Google Scholar]
- 35.Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease (vol 12, pg 362, 2011). Nat Rev Mol Cell Bio. 2011;12(8). 10.1038/nrm3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu L, Horstmann H, Ng C, Hong W. Regulation of Golgi structure and function by ARF-like protein 1 (Arl1). Journal of Cell Science. 2001;114(Pt 24):4543–55. Epub 2002/01/17. . [DOI] [PubMed] [Google Scholar]
- 37.Torres IL, Rosa-Ferreira C, Munro S. The Arf family G protein Arl1 is required for secretory granule biogenesis in Drosophila. Journal of Cell Science. 2014;127(10):2151–60. 10.1242/jcs.122028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hyttinen JMT, Niittykoski M, Salminen A, Kaarniranta K. Maturation of autophagosomes and endosomes: A key role for Rab7. Bba-Mol Cell Res. 2013;1833(3):503–10. 10.1016/j.bbamcr.2012.11.018 [DOI] [PubMed] [Google Scholar]
- 39.Wynant N, Santos D, Van Wielendaele P, Vanden Broeck J. Scavenger receptor‐mediated endocytosis facilitates RNA interference in the desert locust, Schistocerca gregaria. Insect Molecular Biology. 2014;23(3):320–9. 10.1111/imb.12083 [DOI] [PubMed] [Google Scholar]
- 40.Li XX, Dong XL, Zou C, Zhang HY. Endocytic pathway mediates refractoriness of insect Bactrocera dorsalis to RNA interference. Scientific Reports. 2015;5. doi: ARTN 8700 10.1038/srep08700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao D, Gao XW, Xu JP, Liang X, Li QQ, Yao JX, et al. Clathrin-dependent endocytosis plays a predominant role in cellular uptake of double-stranded RNA in the red flour beetle. Insect Biochem Molec. 2015;60:68–77. 10.1016/j.ibmb.2015.03.009 [DOI] [PubMed] [Google Scholar]
- 42.Cappelle K, de Oliveira C, Van Eynde B, Christiaens O, Smagghe G. The involvement of clathrin‐mediated endocytosis and two Sid‐1‐like transmembrane proteins in double‐stranded RNA uptake in the Colorado potato beetle midgut. Insect Molecular Biology. 2016;25(3):315–23. 10.1111/imb.12222 [DOI] [PubMed] [Google Scholar]
- 43.Yoon JS, Shukla JN, Gong ZJ, Mogilicherla K, Palli SR. RNA interference in the Colorado potato beetle, Leptinotarsa decemlineata: Identification of key contributors. Insect Biochem Molec. 2016;78:78–88. 10.1016/j.ibmb.2016.09.002 [DOI] [PubMed] [Google Scholar]
- 44.Branson TF, Jackson JJ. An improved diet for adult Diabrotica virgifera virgifera (Coleoptera, Chrysomelidae). J Kansas Entomol Soc. 1988;61(3):353–5. [Google Scholar]
- 45.Eyun SI, Wang HC, Pauchet Y, Ffrench-Constant RH, Benson AK, Valencia-Jimenez A, et al. Molecular Evolution of Glycoside Hydrolase Genes in the Western Corn Rootworm (Diabrotica virgifera virgifera). PloS one. 2014;9(4). doi: ARTN e94052 10.1371/journal.pone.0094052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23(10):1289–91. 10.1093/bioinformatics/btm091 [DOI] [PubMed] [Google Scholar]
- 47.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40(15). doi: ARTN e115 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Velez AM, Khajuria C, Wang H, Narva KE, Siegfried BD. Knockdown of RNA Interference Pathway Genes in Western Corn Rootworms (Diabrotica virgifera virgifera Le Conte) Demonstrates a Possible Mechanism of Resistance to Lethal dsRNA. PloS one. 2016;11(6). doi: ARTN e0157520 10.1371/journal.pone.0157520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, Kramer KJ. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(32):11337–42. 10.1073/pnas.0504982102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(Delta Delta CT) method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 51.Rodrigues TB, Khajuria C, Wang H, Matz N, Cunha Cardoso D, Valicente FH, et al. Validation of reference housekeeping genes for gene expression studies in western corn rootworm (Diabrotica virgifera virgifera). PloS one. 2014;9(10):e109825 10.1371/journal.pone.0109825 ; PubMed Central PMCID: PMCPMC4214676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.SAS-Institute. SAS User's Manual, Version 9.3. Cary, NC2011.
- 53.Allan AK, Du J, Davies SA, Dow JA. Genome-wide survey of V-ATPase genes in Drosophila reveals a conserved renal phenotype for lethal alleles. Physiol Genomics. 2005;22(2):128–38. Epub 2005/04/28. 10.1152/physiolgenomics.00233.2004 [DOI] [PubMed] [Google Scholar]
- 54.Ren D, Cai Z, Song J, Wu Z, Zhou S. dsRNA uptake and persistence account for tissue-dependent susceptibility to RNA interference in the migratory locust, Locusta migratoria. Insect Molecular Biology. 2014;23(2):175–84. 10.1111/imb.12074 [DOI] [PubMed] [Google Scholar]
- 55.Lee J, Lee J, Ju BG. Drosophila arf72A acts as an essential regulator of endoplasmic reticulum quality control and suppresses autosomal-dominant retinopathy. Int J Biochem Cell B. 2011;43(9):1392–401. 10.1016/j.biocel.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 56.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. Journal of Cell Science. 2010;123(8):1183–9. 10.1242/jcs.066399 [DOI] [PubMed] [Google Scholar]
- 57.Shukla JN, Kalsi M, Sethi A, Narva KE, Fishilevich E, Singh S, et al. Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA biology. 2016;13(7):656–69. Epub 2016/06/02. 10.1080/15476286.2016.1191728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenwald AG, Rhodes MA, Van Valkenburgh H, Palanivel V, Chapman G, Boman A, et al. ARL1 and membrane traffic in Saccharomyces cerevisiae. Yeast. 2002;19(12):1039–56. 10.1002/yea.897 [DOI] [PubMed] [Google Scholar]
- 59.Aronstein K, Pankiw T, Saldivar E. SID-I is implicated in systemic gene silencing in the honey bee. Journal of Apicultural Research. 2006;45(1):20–4. [Google Scholar]
- 60.Bansal R, Michel AP. Core RNAi machinery and Sid1, a component for systemic RNAi, in the hemipteran Iisect, Aphis glycines. Int J Mol Sci. 2013;14(2):3786–801. 10.3390/ijms14023786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong Y, Friedrich M. Nymphal RNAi: systemic RNAi mediated gene knockdown in juvenile grasshopper. Bmc Biotechnol. 2005;5. doi: Artn 25 10.1186/1472-6750-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Firmino AAP, Fonseca FCD, de Macedo LLP, Coelho RR, de Souza JDA, Togawa RC, et al. Transcriptome Analysis in Cotton Boll Weevil (Anthonomus grandis) and RNA Interference in Insect Pests. PloS one. 2013;8(12). doi: UNSP e85079 10.1371/journal.pone.0085079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghosh S, Kakumani PK, Kumar A, Malhotra P, Mukherjee SK, Bhatnagar RK. Genome wide screening of RNAi factors of Sf21 cells reveal several novel pathway associated proteins. Bmc Genomics. 2014;15. doi: Artn 775 10.1186/1471-2164-15-775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gong L, Wang Z, Wang HD, Qi JW, Hu MY, Hu QB. Core RNAi Machinery and Three Sid-1 Related Genes in Spodoptera litura (Fabricius). Int J Agric Biol. 2015;17(5):937–44. [Google Scholar]
- 65.Tian HG, Peng H, Yao Q, Chen HX, Xie Q, Tang B, et al. Developmental Control of a Lepidopteran Pest Spodoptera exigua by Ingestion of Bacteria Expressing dsRNA of a Non-Midgut Gene. PloS one. 2009;4(7). doi: ARTN e6225 10.1371/journal.pone.0006225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu HJ, Chen T, Ma XF, Xue J, Pan PL, Zhang XC, et al. Genome-wide screening for components of small interfering RNA (siRNA) and micro-RNA (miRNA) pathways in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Molecular Biology. 2013;22(6):635–47. 10.1111/imb.12051 [DOI] [PubMed] [Google Scholar]
- 67.Xu WN, Han ZJ. Cloning and phylogenetic analysis of sid-1-like genes from aphids. Journal of Insect Science. 2008;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luo Y, Wang XH, Yu D, Kang L. The SID-1 double-stranded RNA transporter is not required for systemic RNAi in the migratory locust. RNA biology. 2012;9(5):663–71. 10.4161/rna.19986 PubMed PMID: WOS:000306528800018. [DOI] [PubMed] [Google Scholar]
- 69.Wang HD, Gong L, Qi JW, Hu MY, Zhong GH, Gong L. Molecular cloning and characterization of a SID-1-like gene in Plutella xylostella. Arch Insect Biochem. 2014;87(3):164–76. 10.1002/arch.21189 [DOI] [PubMed] [Google Scholar]
- 70.Whangbo JS, Weisman AS, Chae J, Hunter CP. SID-1 Domains Important for dsRNA Import in Caenorhabditis elegans. G3-Genes Genom Genet. 2017;7(12):3887–99. 10.1534/g3.117.300308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valdes VJ, Athie A, Salinas LS, Navarro RE, Vaca L. CUP-1 Is a Novel Protein Involved in Dietary Cholesterol Uptake in Caenorhabditis elegans. PloS one. 2012;7(3). doi: ARTN e33962 10.1371/journal.pone.0033962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoon JS, Gurusamy D, Palli SR. Accumulation of dsRNA in endosomes contributes to inefficient RNA interference in the fall armyworm, Spodoptera frugiperda. Insect Biochem Mol Biol. 2017;90:53–60. Epub 2017/09/28. 10.1016/j.ibmb.2017.09.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.