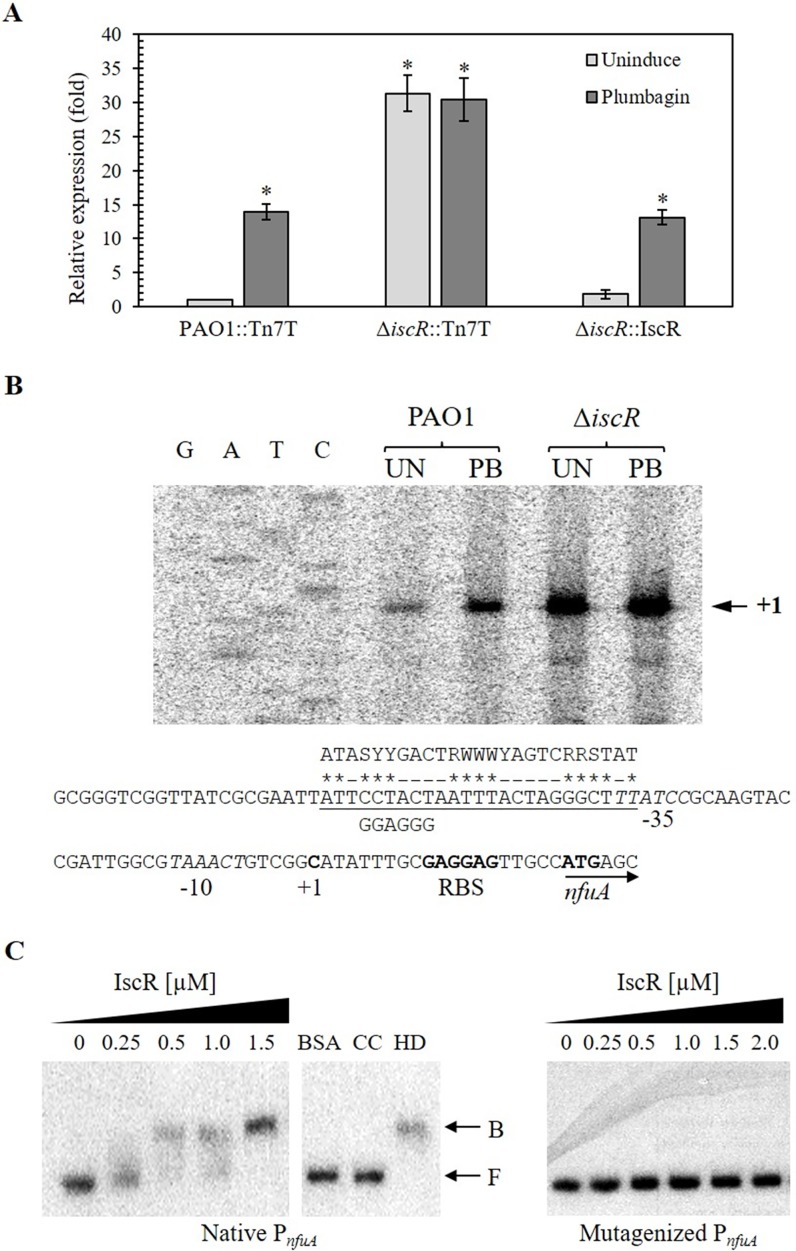
Fig 2. IscR-regulated nfuA expression and nfuA promoter analysis.
(A) IscR-regulated nfuA expression. RNA samples were isolated from uninduced and 0.5 mM plumbagin (PB)-induced cultures of the indicated P. aeruginosa strains. qRT-PCR using primers BT2841 and BT2860 for monitoring nfuA expression and performed as described in the Materials and Methods. Relative expression (fold) is defined as the changes in the nfuA expression levels across multiple samples relative to the level of the uninduced culture of PAO1. The data are presented as the means ± SD from three independent experiments. (B) The primer extension assay was performed using 32P-labeled primer BT3577 and RNA extracted from P. aeruginosa PAO1 and ΔiscR grown under uninduced (UN) and 0.25 mM PB-induced conditions. G, A, T, and C represent the DNA ladder sequence prepared using 32P-labeled primer BT3577 and plasmid pPnfuA as the template. The arrowhead indicates the transcription start site (+1). The -10 and -35 elements are in italic type. The consensus sequence of the Type-I E. coli IscR-binding site is aligned above the corresponding underlined sequence, and the homologous bases are marked with asterisks. The mutated IscR-binding site on the nfuA promoter was aligned below the underlined sequence line. The putative ribosome-binding site (RBS) is indicated in bold type. (C) The electrophoretic mobility shift assay was performed using 32P-labeled native or mutagenized nfuA promoter fragments and increasing concentrations of purified IscR. CC and HD represent an addition of 1 μg unlabeled nfuA promoter and 2.5 μg of heterologous DNA (pUC18 plasmid), respectively, to the binding reaction mixtures containing 3.0 μM IscR. F and B indicate free and bound probes, respectively.