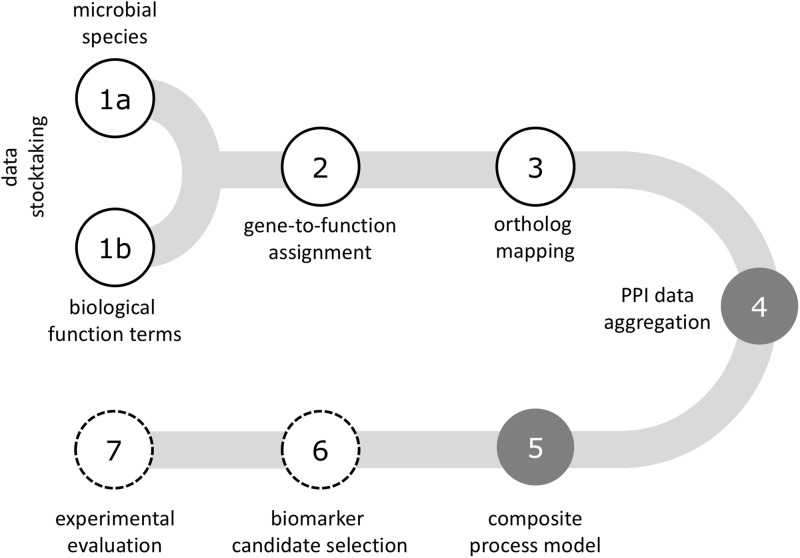
Fig 1. Workflow for candidate biomarker selection.
Microbial species and molecular feature details: step 1—data stocktaking at the microbial species and biological function level; step 2—assignment of genes to biological function terms. Interaction data and networks: step 3—all-against-all ortholog mapping; step 4—aggregation of protein interaction data; step 5—deriving a molecular process model. Biomarker candidates and experimental evaluation: step 6—selection of biomarker candidates utilizing network topology characteristics; step 7—experimental evaluation of biomarker candidates.