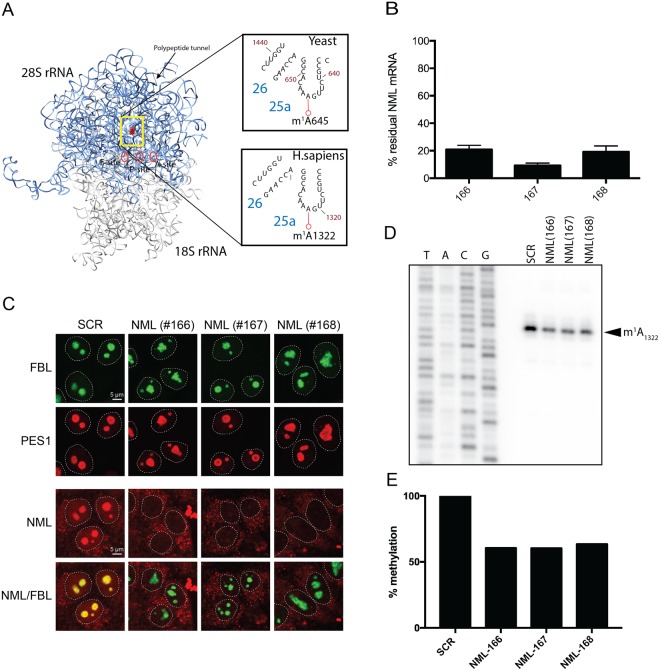
Figure 3.
Validation of nucleomethylin (NML) as the human m1A large subunit rRNA methyltransferase. (A) 3-D structure model of the human rRNA. 4UG0 pdb file together with UCSF-Chimera were used to create the rRNA surface model. The N1-methylated residue A1322 in the 28S rRNA is highlighted as a red sphere. For comparison, the corresponding region of the yeast 25S rRNA is shown in an inset. (B) siRNA-mediated depletion of NML is highly efficient at mRNA level. Total RNA extracted from HCT116 cells transfected with a siRNA specific for NML (siRNA #166 to #168) and incubated for 72 h, was tested by RT-qPCR. (C) NML is a nucleolar protein. Immunofluorescence with an antibody against NML was performed in HCT116 cells stably expressing a green fluorescent fibrillarin construct, which labels the dense fibrillar component of the nucleolus. The granular component protein PES1 was detected with a specific antibody (Bethyl Lab Inc). (D) Primer extension analysis of m1A methylation after siRNA-mediated depletion of NML in HCT116 cells. 32P-labeled primer complementary to nucleotides 1362 to 1381 of human 28S rRNA were used for methylation analysis of m1A1322. (E) The reduction of stop intensity at the m1A1322 in siRNA-mediated depleted cells was quantified using ImageJ software and estimated to be of ~60%.