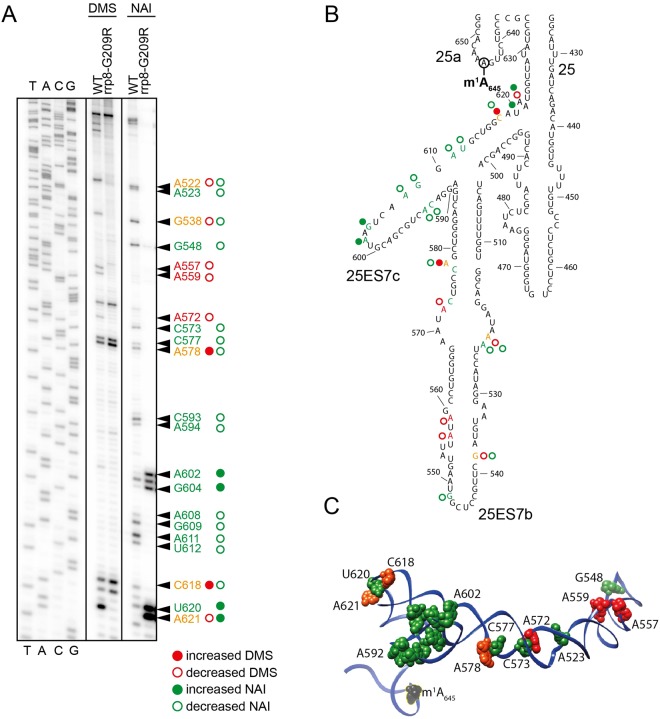
Figure 4.
Loss of m1A645 leads to structural changes within the LSU. (A) Representative gel showing structure probing with DMS or SHAPE analysis with NAI in the rrp8G209R loss of methylation mutant. 32P-labeled primer (helix25_StrPrb) complementary to nucleotides 2428 to 2448 of human 28S rRNA were used for the analysis. Bands corresponding to modified residues are marked in red for DMS, green for NAI and orange for both. (B) 2-D structure model with mapped modified residues as described in (A). (C) The residues sensitive to chemical modification are represented as spheres on a 3-D structure model of helix 25 (color scheme as in panel A). The model was made using Chimera and PDB file 4V88.