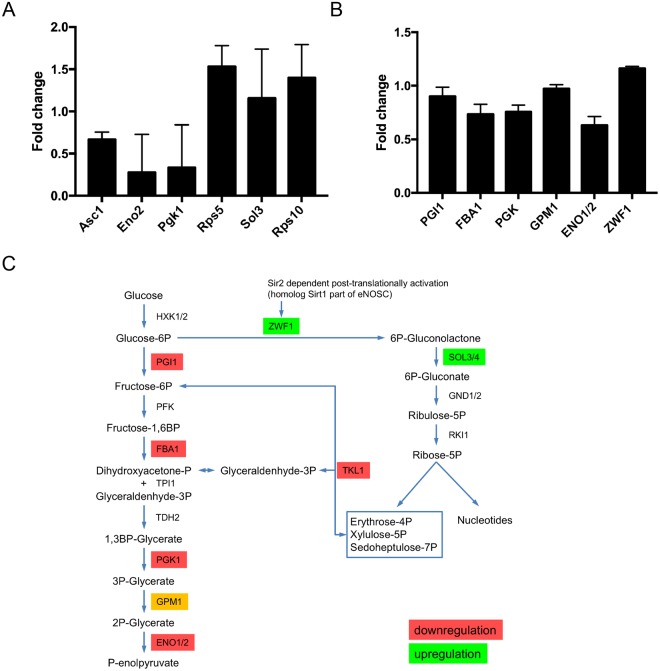
Figure 7.
Quantitative PCR and biochemical activities of differentially regulated proteins in Δrrp8. (A) RTqPCR analysis of mRNAs from proteins that were differentially regulated in the rrp8 mutant strain are represented as fold change in mRNA expression. Expression of Asc1, Rps5, Eno2 and Pgk1 is regulated at the transcriptional level, whereas mRNA levels of Sol3 and Rps10A are not different from their wild-type level of expression. Data analyzed with the software REST®.41. (B) Regulation of specific activities of enzymes of the carbohydrate metabolism within Δrrp8. Changes in the specific enzyme activities of phosphoglucose isomerase (PGI1), fructose-1,6-bisphosphate aldolase (FBA1), phosphoglycerate kinase (PGK1), phosphoglycerate mutase (GPM1), enolase (ENO1/ENO2) and glucose-6-phosphate dehydrogenase (ZWF1) are represented as fold change. Activities of enzymes involved in the glycolytic pathway are downregulated due to deletion of RRP8 as compared to wild-type. Only activity of ZFW1, first enzyme of the pentose-phosphate pathway, is slightly upregulated. (C) Schematic overview of regulations in the carbohydrate metabolism in cells with inactive RRP8.