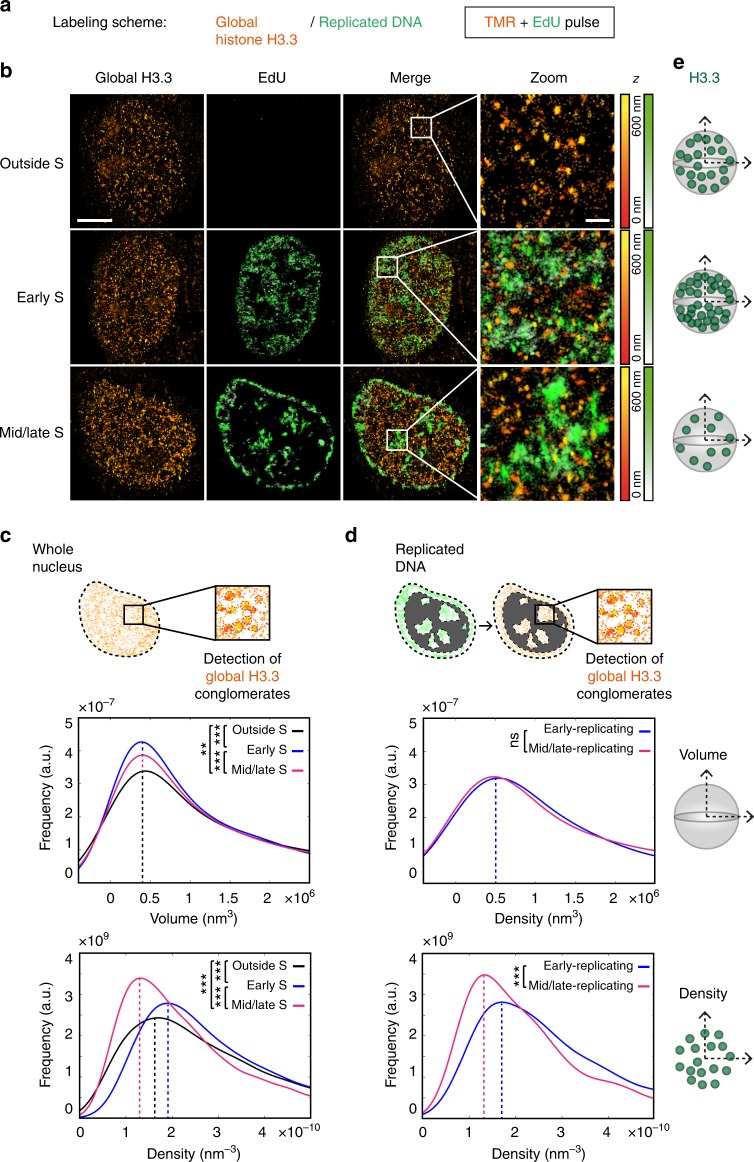
Fig. 3.
Global distribution of H3.3 throughout S phase using STORM assay. a Labeling scheme using H3.3-SNAP to follow global H3.3 as described in Fig. 2. b Representative STORM images of global H3.3 (TMR, orange) and replicated DNA (EdU, green) in HeLa H3.3-SNAP. EdU labeling, as described in Fig. 2, allows the selection of cells outside S phase, in early S phase and mid/late S phase. The color gradient corresponds to the z range. Scale bars represent 5 μm. Insets represent enlarged images of selected area where scale bars correspond to 600 nm. c For the H3.3-enriched areas—defined as conglomerates—in the whole nucleus, the plots show the distribution of volume (top) or density (bottom) of H3.3 conglomerates in cells outside S phase (black), in early S phase (blue) and in mid/late S phase (magenta). For c, d, N = 11, 8 and 10 cells for outside S phase, early S phase and mid/late S phase respectively. The p values (using Mann–Whitney–Wilcoxon test): ∗∗∗p ≤ 0.001; ∗∗p ≤ 0.01; ns not significant. d For H3.3 conglomerates in regions of replicated DNA, the plots show the distribution of volume (top) or density (bottom) of H3.3 conglomerates in cells in early S phase (blue) and in mid/late S phase (magenta). e Scheme summarizing the changes in volume and density of H3.3 conglomerates outside S phase, in early S phase and in mid/late S phase