Abstract
Background
Human infections with the H7N9 virus could lead to lung damage and even multiple organ failure, which is closely associated with a high mortality rate. However, the metabolic basis of such systemic alterations remains unknown.
Methods
This study included hospitalized patients (n = 4) with laboratory-confirmed H7N9 infection, healthy controls (n = 9), and two disease control groups comprising patients with pneumonia (n = 9) and patients with pneumonia who received steroid treatment (n = 10). One H7N9-infected patient underwent lung biopsy for histopathological analysis and expression analysis of genes associated with lung homeostasis. H7N9-induced systemic alterations were investigated using metabolomic analysis of sera collected from the four patients by using ultra-performance liquid chromatography-mass spectrometry. Chest digital radiography and laboratory tests were also conducted.
Findings
Two of the four patients did not survive the clinical treatments with antiviral medication, steroids, and oxygen therapy. Biopsy revealed disrupted expression of genes associated with lung epithelial integrity. Histopathological analysis demonstrated severe lung inflammation after H7N9 infection. Metabolomic analysis indicated that fatty acid metabolism may be inhibited during H7N9 infection. Serum levels of palmitic acid, erucic acid, and phytal may negatively correlate with the extent of lung inflammation after H7N9 infection. The changes in fatty acid levels may not be due to steroid treatment or pneumonia.
Interpretation
Altered structural and secretory properties of the lung epithelium may be associated with the severity of H7N9-infection-induced lung disease. Moreover, fatty acid metabolism level may predict a fatal outcome after H7N9 virus infection.
Keywords: H7N9 virus, Lung epithelium, Fatty acid metabolism, Palmitic acid, Metabolomics
Research in context.
Evidence before this study
Since its emergence in 2013, the novel avian-origin influenza A H7N9 virus has caused seasonal epidemics every year. A more highly pathogenic H7N9 variant has been identified recently. This variant could increase the potential threat of a greater pandemic, especially if it acquires efficient human-to-human transmissibility. Infections with the H7N9 virus may lead to lung damage and even multiple organ failure, which is closely associated with a high mortality rate. No previous study has assessed the systemic alterations associated with H7N9 virus infection.
Added Value of This Study
In the present study, we observed that the H7N9 virus may cause the disruption of epithelial structure and the alteration of epithelial secretory properties of the lung. The virus elicits systemic alterations that may also contribute to virus-induced multiple organ injuries. Metabolomic analysis of serum samples from H7N9-infected patients revealed that fatty acid metabolism may be repressed after H7N9 infection. Serum levels of palmitic acid, erucic acid, and phytal may negatively correlate with disease severity after H7N9 infection.
Implications of All the Available Evidence
To our knowledge, this is the first study to analyze the systemic alterations of serum metabolites in H7N9-infected patients. Serum levels of palmitic acid, erucic acid, and phytal may predict a fatal outcome after H7N9 infection. Targeting epithelial regeneration in the lung may be a future option for treating H7N9 infections.
1. Introduction
The emergence in 2013 of the novel avian-origin influenza A H7N9 virus imposed a great challenge not only to poultry production but also to public health because of the high rates of morbidity and mortality. As of February 2018, H7N9 viruses have caused five seasonal epidemic waves, with the number of laboratory-confirmed human cases increasing appreciably [52]. Travel-related cases have been confirmed outside China, including Canada and Malaysia [26]. Although the occurrence of international spread has not yet been indicated, recent evidence demonstrates that a more highly pathogenic H7N9 variant has evolved, thereby increasing the potential threat of a greater pandemic, especially if it acquires efficient human-to-human transmissibility (Bertran et al., [1]; [28, 54]). Close monitoring of the virological and molecular characteristics of H7N9 viruses is therefore urgently needed. Moreover, clinical severity assessments of H7N9-infected patients are crucial to develop more effective control measures.
In patients with poultry contact, H7N9 virus infection may result in critical illnesses, including rapidly progressing pneumonia, respiratory failure, and acute respiratory distress syndrome (ARDS). Therefore, the administration of steroids, including methylprednisolone, is common in the intensive care unit (ICU) to limit lung inflammation [35, 37]. Hypoxic respiratory failure was reported as the leading cause of death [3]. As a result, endotracheal intubation and protective mechanical ventilation (MV) have become the mainstay of supportive therapy in the ICU [17, 38]. Severe ARDS associated with H7N9 virus infection leads to the use of extracorporeal membrane oxygenation (ECMO), originally applied to support the respiratory function of pediatric patients [24, 36]. Nevertheless, approximately 40% of patients do not survive these treatments, possibly because of the following three factors. First, the clinical prognosis of H7N9-infected patients is closely related to underlying medical complications [51]. Second, the severity is determined by the time interval from disease onset to antiviral treatment. Lastly, the mortality increases when H7N9 virus infection induces multiple organ failure [56].
The key cause of systemic alterations in H7N9 infection is lung damage characterized by impaired mucociliary clearance and disturbed alveolar homeostasis. As regenerating machines, both club and alveolar type 2 epithelial cells are targets of H7N9 viruses, resulting in the inefficient restoration of airway barriers and overproduction of mucus and surfactants [48]. Thus, hospitalized patients confirmed to have H7N9 virus infection are immediately started on antiviral therapy using existing neuraminidase inhibitors, such as oseltamivir and zanamivir, or neutralizing antibodies against hemagglutinin [19, 22, 24, 57]. As the H7N9 virus travels via the circulatory system to target more cell types (Hu et al., 2015), it is hypothesized to elicit systemic alterations that may also contribute to virus-induced multiple organ injuries.
Metabolomics, a rapidly emerging field of “omics” research, presents pathobiological profiles that encompass both microbial and host interactions [42, 45]. Metabolomic approaches have identified novel biomarkers and new pathobiological pathways associated with virus infections [9, 47]. However, to date, no study has applied a metabolomic approach in H7N9-infected patients.
To address this, we used serum samples from four patients who were hospitalized because of H7N9 infection to profile their serum metabolome. We found that the H7N9-infected patients had metabolic disorders possibly associated with fatty acid metabolism. To determine its possible relationship with disease severity, we tried to correlate the abundance of identified serum metabolites with the extent of lung inflammation evaluated using chest digital radiography and absolute serum lymphocyte counts. A three-metabolite set may be associated with the resolution of lung inflammation during H7N9 infection. We also observed that severe inflammation induced by H7N9 causes the altered expression of airway mucin and alveolar surfactants.
2. Materials and Methods
2.1. Ethics Statement
The histopathological features were reported for one patient whose relative consented to limited postmortem biopsy. Full autopsy of the patient could not be performed because of religious and other reasons. The institutional review board of Tianjin Haihe Hospital provided written approval and judged that all methods used in the study met relevant ethical guidelines and regulations (ethical # 2016HHKT-01).
2.2. Subjects
Four H7N9-infected patients were admitted to Tianjin Haihe Hospital between June 13, 2016 and June 20, 2017. During this period, nine consenting healthy volunteers, nine patients with pneumonia (according to the 2007 American Thoracic Society Inclusion Criteria), and ten patients with pneumonia who received steroid treatment were recruited as controls from the same hospital. General participant information, including age, sex, recent poultry exposure, disease duration, and clinical treatments and medication, was collected using a standard form. As described previously [12], for the diagnosis of H7N9 infection, RNA was extracted from throat swabs and sputum of the patients, and a multiplex one-step real-time PCR assay was developed to detect the H7N9 virus by using primers targeting the conserved M and RNase P genes, as well as the hemagglutinin and neuraminidase genes of the H7N9 virus. Serum samples were collected between 6:00 and 8:00 a.m. from all patients, and one part was used for routine clinical measurements and the other part was immediately stored in a −80°C refrigerator for further metabolomic analysis. Tru-cut postmortem biopsy specimens were obtained from the left lung at the sixth intercostal space posteriorly within 18–19 h after death. Two successful biopsies were conducted aseptically according to standard guidelines (Otto et al., 2015), with one specimen being put into a cell lysis medium for gene expression analysis using quantitative PCR and the other into 4% formalin for histopathological examination. For comparison, normal lung tissue adjacent to the tumor was also surgically removed from patients with lung adenocarcinoma.
2.3. Hematoxylin and Eosin Staining
Lung tissues were fixed in 10% neutral-buffered formalin, and then embedded in paraffin. Five-micrometer sections were collected and stained with hematoxylin and eosin (H&E) solution (Solarbio Science & Technology Co, Ltd., Beijing, China). Images were captured using an Olympus IX71 microscope with a DP80 camera (Olympus Corporation, Tokyo, Japan).
2.4. Scanning Electron Microscopy
Lung biopsy specimens were fixed in 2.5% glutaraldehyde for 2 h, and then immobilized in 2% osmium tetroxide. In brief, the tissues were dehydrated through sequential washes in 50, 70, 90, 95, and 100% ethanol followed by immersion in EPON812 (Simga Aldrich, St. Louis, MO, USA). Ultrathin sections were collected onto copper grids and then counterstained with uranyl acetate and lead citrate. Images were acquired using a FEI Tecnai Spirit transmission electron microscope (ThermoFisher Scientific, Hillsboro, Oregon, USA).
2.5. Quantitative Real-Time PCR
RNA was extracted from lung biopsy specimens and normal lung tissue by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The whole RNA was reverse transcribed using oligo (dT) primers for 1 h at 50 °C using the TIANScript RT kit (Tiangen Biotech, Co., Ltd., Beijing, China) according to the manufacturer's protocol. Quantitative PCR analysis was performed using the SYBR Green method. Specific gene primers were designed using the Primer-Quest SM software (http://sg.idtdna.com/PrimerQuest/Home/Index), and then commercially produced (BGI Tech, Shenzhen, China; listed in Table S1). The cDNA amplification reactions were performed using a Light Cycler 96 real-time PCR system (Roche Diagnostics, Indianapolis, IN, USA) with the following reaction conditions: an initial heating cycle of 95 °C for 2 min, and 40 cycles of denaturation at 95 °C for 25 s, primer annealing at 60 °C for 25 s, and extension at 72 °C for 20 s. Melting curves were used for clarifying the identity of amplicons, and the housekeeping gene β-actin was used as an internal control. The relative mRNA expression levels of targeted genes were evaluated using the comparative Cq (quantification cycle) method normalized to the expression level of β-actin mRNA in the same sample.
2.6. Metabolite Extraction
Before metabolomic analysis, the samples were defrosted at room temperature for <20 min. Briefly, 400 μL of acetonitrile was added to the samples in a 4:1 (v/v) ratio. These sample mixtures were then homogenized by shaking them for 30 s and centrifuged at 15,000 ×g for 20 min at 4 °C. A 400-μL aliquot of the supernatant was pipetted into a 5-mm test tube and then loaded into the ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) system for analysis.
2.7. Metabolome Analysis
Samples were measured using a nano LC system (Thermo Fisher Scientific, Germering, Germany) coupled on-line to a hybrid linear ion-trap/Orbitrap™ mass spectrometer (LTQ-Orbitrap-XL, Thermo Fisher Scientific, Bremen, Germany). Samples were loaded onto a trap column (PepMap C18; 300-μm internal diameter [ID], 5-mm length, 5-μm particle size, 100-Å pore size; Thermo Fisher Scientific), and then washed and desalted for 15 min by using 0.1% trifluoroacetic acid in water as the loading solvent. The trap column was then switched in-line with the analytical column (PepMap C18; 75-μm ID, 250-mm length, 3-μm particle size, 100-Å pore size; Thermo Fisher Scientific) and peptides were eluted using the following binary gradient: starting with 100% solvent A and B, where solvent A consisted of 2% acetonitrile and 0.1% formic acid in water, and solvent B consisted of 80% acetonitrile and 0.08% formic acid in water. The column flow rate was set at 200 nL/min. For electro-spray ionization (ESI), nano ESI emitters (New Objective, Woburn, MA, USA) were used and a spray voltage of 4.5 kV was applied. For MS detection, a data-dependent acquisition method was used: high-resolution survey scan from 50 to 1000 (m\z). Orbitrap full-scan spectra and ion-trap MS/MS fragmentation spectra were acquired partially simultaneously.
2.8. Compound Identification, Quantification, and Data Curation
The UPLC-MS raw data were converted and processed using MZmine 2 (http://mzmine.sourceforge.net/). The peaks were aligned and normalized to the sum of all the detected peaks. Briefly, chromatograms were built and peaks were recognized using the local-minimum search function; thereafter, the ion intensities, matching m/z, and retention time were grouped into peak lists. The online Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.kegg.jp/) classifications and human metabolome database (HMDB, http://www.hmdb.ca/) were used to align the molecular mass data (m/z) to identify the metabolites. The processed and normalized data were imported into SIMCA-P (Umetrics, Umea, Sweden) for multivariate statistical analysis. To distinguish H7N9 from the controls, the orthogonal projections to latent structure-discriminant analysis (OPLS-DA) model was introduced to determine the maximum separation between different kinds of samples according to the sample classification information. Based on the OPLS-DA model, the specific metabolites were determined by applying the Mann-Whitney U test (SPSS 15.0) with a P-value threshold of 0.05. The names of the metabolites were identified by searching the online HMDB database via aligning the molecular mass data (m/z). Then, the specific metabolites were clustered using hierarchical cluster analysis and visualized using Hemi (Heatmap Illustrator, version 1.0) software (http://hemi.biocuckoo.org/down.php). Multiple logistic regression analysis was carried out and receiver-operating characteristic (ROC) analysis was used to evaluate the diagnostic value of metabolites. The area under the curve and 95% confidence intervals were calculated to determine the specificity and sensitivity for H7N9. Thereafter, the specific metabolites were exported individually and imported into MetaboAnalyst 2.0 (http://nar.oxfordjournals.org/content/early/2012/05/01/nar.gks374.full) for analyzing the metabolic pathway.
2.9. Statistical Analyses
The Mann-Whitney U test was used for multivariate comparisons, and hierarchical cluster analysis was performed using Hemi analysis. The software used for other statistical analysis was SPSS for Windows, Version 15.0 (SPSS Inc., Chicago, IL, USA) unless specifically mentioned otherwise. The threshold for significance was P < 0.05. A combination of univariate and multivariate bioinformatic approaches were employed using the R script-based online tool MetaboAnalyst 2.0, a comprehensive tool suitable for analyzing metabolomic data.
3. Results
3.1. General Treatments and Outcomes of H7N9-Infected Patients
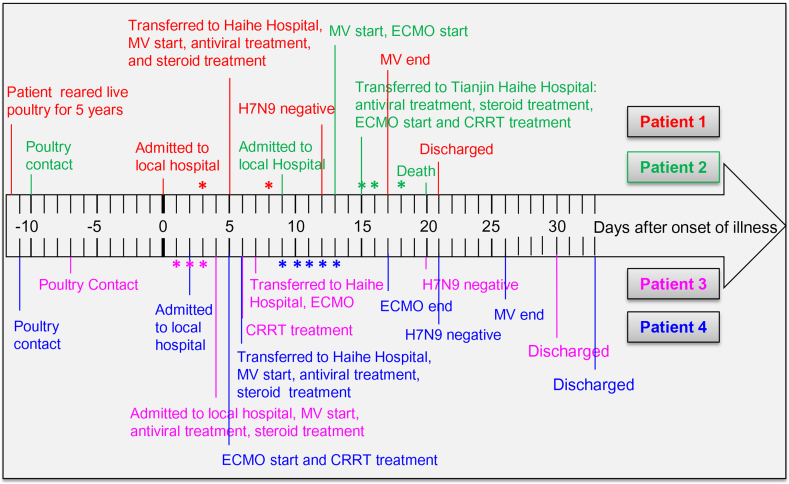
The four elderly patients (two men and two women) included in the study, who were later found to have laboratory-confirmed H7N9 infection, as described previously [12], had a history of poultry contact (Fig. 1). They were admitted to hospital with high fever, cough, chest distress, and dyspnea (Table 1, Table 2). They also had septic shock and developed ARDS. Therefore, they were immediately administered antiviral medicine, including oseltamivir and peramivir, together with methylprednisolone and immunoglobulin intravenously (Fig. 1). To control secondary infection, antibiotics including linezolid, imipenem, and cilastatin sodium were administered (Table 1). To improve gas exchange, oxygen therapy was applied to all four patients. Patients 2, 3, and 4 were also treated with ECMO (Fig. 1). These three patients had acute renal damage; therefore, continuous renal-replacement therapy (CRRT) was necessary (Table 1). Patients 1 and 4 had hypertension as the only known underlying condition; they responded to this treatment regimen very well and were cured and discharged after the real-time PCR analysis for the H7N9 virus in the airway lavage fluid and blood tested negative (Fig. 1). In addition to hypertension, patients 2 and 3 had other underlying conditions, including coronary heart disease, and ended up having worse outcomes. Patient 2 died 5 days after admission to Tianjin Haihe Hospital, and 20 days after disease onset (Fig. 1). As requested by her family members, patient 3 was discharged when her airway lavage fluid and blood tested negative for H7N9. Her medical conditions were recovering and stable; however, on follow-up, it was found that she died a few days after discharge. These data indicated that H7N9 may induce severe lung inflammation.
Fig. 1.
Timeline of the clinical course of four patients with confirmed H7N9 infection. The timeline illustrating the clinical course from poultry contact; hospitalization and clinical treatments, such as antiviral treatment, steroid treatment, mechanical ventilation (MV), extracorporeal membrane oxygenation (ECMO), and continuous renal-replacement therapy (CRRT); and treatment outcomes. *represents the time points of serum collection for metabolomic analysis.
Table 1.
Demographic characteristics, complications, treatment, and outcomes.
| Characteristic | Patient 1 | Patient 2 | Patient 3 | Patient 4 |
|---|---|---|---|---|
| Age (years) | 62 | 63 | 67 | 58 |
| Sex | Male | Male | Female | Female |
| Occupation | Farmer | Retired | Retired | Farmer |
| Cardinal symptom | High fever, chest distress, shortness of breath, dyspnea | Cough, high fever, dyspnea, hemoptysis | High fever, chest distress, dyspnea | High fever, chest pain, cough, chest distress, shortness of breath, abdominal pain |
| Underlying conditions | Hypertension | Hypertension, coronary heart disease, COPD, pulmonary heart disease | Hypertension, coronary heart disease, tuberculosis of lymph nodes | Hypertension |
| Poultry exposure in the past 2 weeks | Yes | Yes | Yes | Yes |
| Septic shock | Yes | Yes | Yes | Yes |
| ARDS | Yes | Yes | Yes | Yes |
| Acute renal damage | No | Yes | Yes | Yes |
| Encephalopathy | No | No | Yes | Yes |
| Rhabdomyolysis | No | Yes | No | Yes |
| Secondary infections | No | No | Yes | Yes |
| Oxygen therapy | Yes | Yes | Yes | Yes |
| Extracorporeal membrane oxygenation | No | Yes | Yes | Yes |
| Continuous renal-replacement therapy | No | Yes | Yes | Yes |
| Antibiotic therapy | Linezolid, imipenem, cilastatin sodium, piperacillin, teicoplanin, moxifloxacin | Linezolid, imipenem, cilastatin sodium, vancomycin, piperacillin, teicoplanin | Linezolid, imipenem, cilastatin sodium, vancomycin, piperacillin, teicoplanin | Imipenem and cilastatin, linezolid, cefoperazone and sulbactam, moxifloxacin |
| Antiviral agents | Oseltamivir | Oseltamivir, peramivir | Oseltamivir, peramivir | Oseltamivir |
| Steroids | Methylprednisolone | Methylprednisolone | Methylprednisolone | Methylprednisolone |
| Intravenous immunoglobulin therapy | Yes | Yes | Yes | Yes |
| Duration of hospital stay | 16 days | 7 days | 26 days | 28 days |
| Prognosis | Cured | Death | Discharge when H7N9 was negative | Cured |
Table 2.
General participant characteristics.
| Healthy | Pneumonia | Pneumonia + steroids | H7N9 + steroids | |
|---|---|---|---|---|
| Number of subjects (n) | 9 | 9 | 10 | 4 |
| Age (years) | 53.6 ± 8.1 | 60.7 ± 7.0 | 63.8 ± 9.0 | 60.4 ± 2.9 |
| Sex (F/M) | 5/4 | 5/4 | 5/5 | 2/2 |
| Chronic smoker | 4/9 | 4/9 | 5/10 | 2/4 |
| Duration of hospital stay (days) | / | 14 ± 6.4 | 18.2 ± 10.3 | 24.7 ± 3.9 |
| Fever | 0/9 | 6/9 | 6/10 | 4/4 |
| Cough | 0/9 | 9/9 | 10/10 | 3/4 |
| Hemoptysis | 0/9 | 2/9 | 0/10 | 0/4 |
| Dyspnea | 0/9 | 3/9 | 7/10 | 4/4 |
| Nausea or vomiting | 0/9 | 0/9 | 1/10 | 1/4 |
| Diarrhea | 0/9 | 0/9 | 0/10 | 0/4 |
| Advanced respiratory support1 | 0/9 | 3/9 | 9/10 | 4/4 |
| Shock | 0/9 | 1/9 | 3/10 | 3/4 |
| Respiratory failure | 0/9 | 2/9 | 8/10 | 4/4 |
| Glucocorticoid (total, grams) | 0 | 0 | 453.8 ± 276.32 | 220.0 ± 134.22 |
| Infections | ||||
| Bacterial | / | 6/9 | 8/10 | 0/4 |
| Viral | / | 3/93 | 2/103 | 4/4 |
| Fungal | / | 0/9 | 0/10 | 0/4 |
| Underlying conditions | ||||
| Coronary heart disease | / | 4/9 | 5/10 | 2/4 |
| Chronic gastritis | / | 2/9 | 2/10 | 0/4 |
| Hypertension | / | 4/9 | 2/10 | 4/4 |
| Diabetes | / | 2/9 | 1/10 | 0/4 |
| COPD | / | 2/9 | 4/10 | 1/4 |
Advanced respiratory support includes high flow nasal cannula, and invasive mechanical ventilation.
Methylprednisolone was used.
Infected with influenza B virus.
3.2. Lung Epithelium is Injured During H7N9-Induced Inflammation
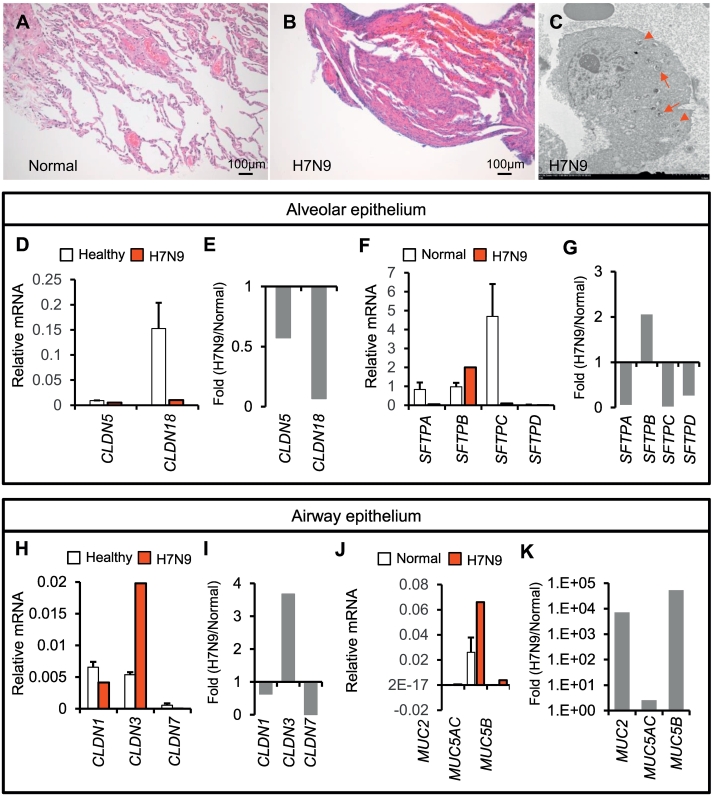
Virus-induced acute lung inflammation is characterized by the infiltration of inflammatory cells, and is usually associated with epithelial injuries, resulting in increased secretion and fluid buildup. Compared to the normal lung tissue adjacent to the lung adenocarcinoma (68-year-old male patient; T1bN0M0; right lower lobe), the biopsied lung tissue from patient 2 with H7N9 infection showed obvious inflammatory infiltrates (Fig. 2A–B). Moreover, this patient showed diffuse alveolar hemorrhage, suggesting severe injuries to the alveolar structure (Fig. 2B). Similar to other influenza virus infections [13, 55], H7N9 virus infection may also induce the formation of autophagosomes that promote the host's capacity of limiting viral infection (Fig. 2C). Quantitative PCR analysis indicated that claudin 5 and 18, which are tight junction genes that maintain alveolar epithelial structure, were downregulated in the H7N9-infected lung (Fig. 2D–E). Moreover, the expression of surfactant protein B (SFTPB) was promoted, while that of SFTPA, SFTPC, and SFTPD was significantly inhibited, in the lung of patient 2 than in the normal lung (Fig. 2F–G). In conducting airways, claudin 3 gene expression was increased, while claudin 1 and 7 gene expressions were reduced (Fig. 2H–I). The expression of mucin genes, including MUC2, MUC5AC, and MUC5B, was significantly increased to a different extent in the infected lung (Fig. 2J–K). These preliminary data suggested that the integrity of both the conducting airway epithelium and alveolar epithelium may be challenged by H7N9 virus infection.
Fig. 2.
Lung epithelial barrier is severely injured during H7N9-induced lung inflammation. (A–B) The biopsied tissues isolated from patient 2 are analyzed using H&E staining of normal lung tissue adjacent to the lung adenocarcinoma (Normal) as the control. (C) Electron microscopic scanning identifies the formation of autophagosomes in alveolar type 2 cells from patient 2 (arrows). Lamellar bodies in alveolar type 2 cells are indicated using arrow heads. (D–G) Expression of claudins and alveolar surfactants is analyzed in the biopsied lung tissue by using quantitative PCR. Relative levels to β-actin and fold changes (H7N9/Normal) are presented separately. (H–K) Expression of airway mucins and claudins is analyzed and presented in the same manner as above.
3.3. H7N9 Infection Impairs Fatty Acid Metabolism
In addition to lung damage, H7N9 infection has been demonstrated to cause multiple organ failure [56], suggesting its pathological effects are systemic. Such systemic alteration induced by the H7N9 virus may be reflected by a change in serum metabolites. To examine this, healthy controls were recruited in the present study (n = 9). Since steroid or pneumonia itself could contribute to the changes in serum metabolites, in order to exclude these effects, we also included two other disease controls, including patients who received steroid treatment (n = 10) and patients who were hospitalized with severe non-H7N9 pneumonia (n = 9). To minimize confounding factors, H7N9-infected patients were compared with age- and sex-matched control subjects in this analysis.
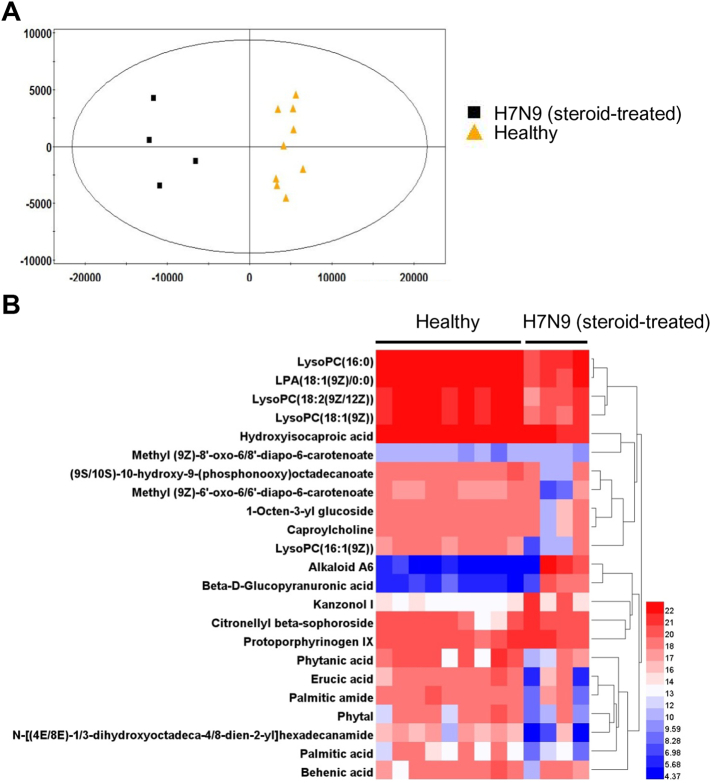
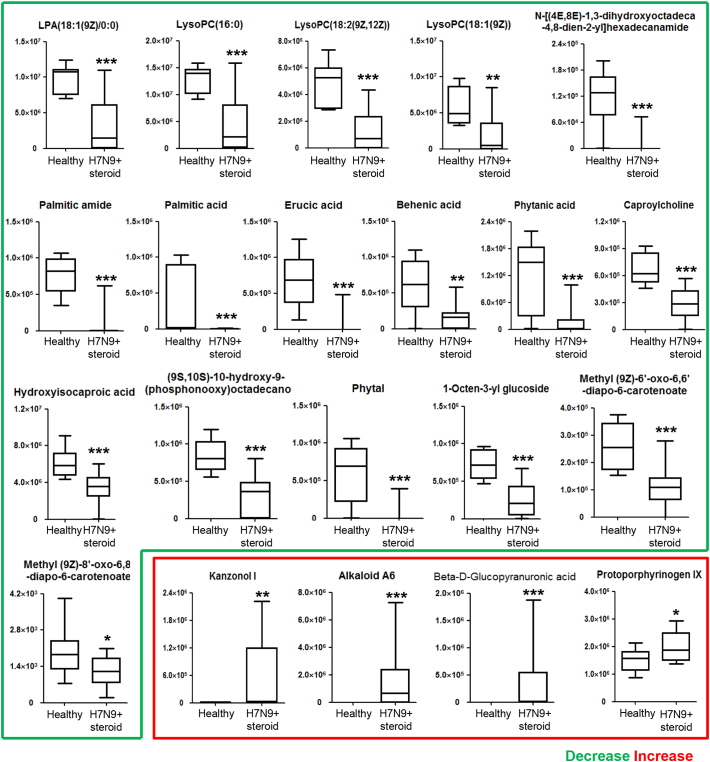
UPLC-MS was adopted in the present study to determine the difference in the serum metabolome of H7N9-infected patients as compared to those in the healthy group and two disease control groups. OPLS-DA of the initial data set obviously separated the H7N9-infected patients receiving steroid treatment (H7N9 + steroid) from the healthy controls (Fig. 3A). The differentially expressed metabolome included 23 metabolites (Fig. 3B). Two of them were excluded after validating the significance of changes by using the nonparametric univariate method (Mann-Whitney-Wilcoxon test) (Fig. 4). The remaining 21 metabolites included four upregulated metabolites, namely, kanzonol I, alkaloid A6, protoporphyrinogen IX, and beta-d-glucopyranuronic acid, in the serum samples of H7N9-infected patients rather than in those from the healthy group (Fig. 4). More metabolites were downregulated, including LysoPC(16:0), LPA(18:1(9Z)/0:0), LysoPC(18:2(9Z,12Z)), LysoPC(18:1(9Z)), palmitic acid, hydroxyisocaproic acid, phytanic acid, erucic acid, behenic acid, palmitic amide, phytal, caproylcholine, (9S,10S)-10-hydroxy-9-(phosphonooxy) octadecanoate, 1-octen-3-yl glucoside, N-[(4E,8E)-1,3-dihydroxyoctadeca-4,8-dien-2-yl] hexadecanamide, methyl (9Z)-8′-oxo-6,8′-diapo-6-carotenoate, and methyl (9Z)-6′-oxo-6,6′-diapo-6-carotenoate (Fig. 4).
Fig. 3.
Serum metabolomic profile of H7N9-infected patients as determined by UPLC-MS. (A) Supervised (OPLS-DA) score scatter plot of H7N9-infected patients receiving steroid treatment (H7N9 + steroid) and healthy controls (healthy) from the UPLC-MS data. (B) Heat map of significantly altered metabolites detected by UPLC-MS. Hierarchical clustering of the 23 metabolites is shown between the healthy group (n = 9) and H7N9 + steroid group (n = 4) in the discovery analysis.
Fig. 4.
Differential metabolites in the sera of H7N9-infected patients. Supervised univariate nonparametric analysis (Mann-Whitney U test) is used to confirm 21 serum metabolites out of 23 that distinguish H7N9-infected patients from healthy controls. Boxes represent the interquartile ranges (IQRs) between the first and third quartiles, and the lines inside the boxes represent the median; whiskers represent the lowest or highest values within 1.5 times the IQR from the first or third quartiles. *P < 0.05, **P < 0.01, ***P < 0.001.
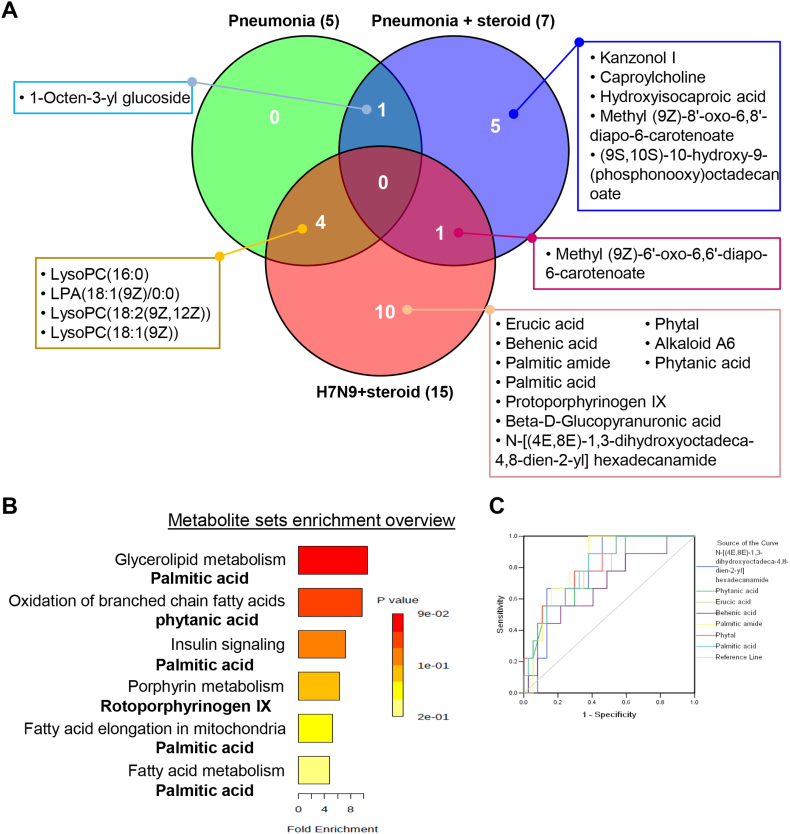
Venn diagram analysis of these 21 serum metabolites indicated that steroid treatment could affect the abundance of kanzonol I, caproylcholine, hydroxyisocaproic acid, methyl (9Z)-8′-oxo-6,8′-diapo-6-carotenoate, and (9S,10S)-10-hydroxy-9-(phosphonooxy) octadecanoate in the serum (Fig. 5A). Besides kanzonol I and (9S,10S)-10-hydroxy-9-(phosphonooxy) octadecanoate, many more metabolites can be altered by steroid treatment, as evidenced by the direct metabolomic analysis of serum samples from patients with severe pneumonia and patients with pneumonia undergoing steroid treatment (Fig. S1). 1-Octen-3-yl glucoside may be related to a non-H7N9 pneumonia condition (Fig. 5A). Direct metabolomic analysis of serum samples from healthy controls and patients with severe pneumonia revealed that many more metabolites can be altered during pneumonia, including LysoPC(16:0), LysoPC(18:0), LysoPC(20:3(5Z/8Z/11Z)), and LysoPC(24:0) (Fig. S2). However, H7N9-infection-associated pneumonia could also lead to changes in the serum abundance of LysoPC(16:0), LPA(18:1(9Z)/0:0), LysoPC(18:2(9Z,12Z)), and LysoPC(18:1(9Z)) (Fig. 5A). Additionally, ten potential serum metabolites were identified to be related to H7N9 virus infection, including palmitic acid, erucic acid, and phytal (Fig. 5A). To investigate the potential metabolic disruptions in H7N9-infected patients, these ten metabolites were prioritized when conducting the metabolic pathway analysis. We observed that H7N9 virus may lead to alterations in insulin signaling, porphyrin metabolism, and, more likely, fatty acid metabolism (Fig. 5B). ROC analysis confirmed those associations and helped evaluate the possible diagnostic value of these metabolites (Fig. 5C).
Fig. 5.
H7N9 infection alters serum fatty acid metabolism. (A) Venn diagram analysis of pneumonia, pneumonia with steroid treatment, and H7N9 infection with steroid treatment groups identifies ten potential serum metabolites specific to H7N9 infection. (B) Metabolite enrichment analysis indicates that the metabolites that distinguish H7N9 infection are related to lipid metabolism. (C) Receiver-operating characteristic curves (ROC curves) showing the performance of models including the metabolites that distinguish H7N9-infected patients from healthy controls and disease controls (pneumonia and pneumonia with steroid treatment groups). AUC indicates the area under the curve, and CI indicates the confidence interval.
3.4. Potential Relevance of the Three-Metabolite Set to the Prognosis of H7N9 Infection
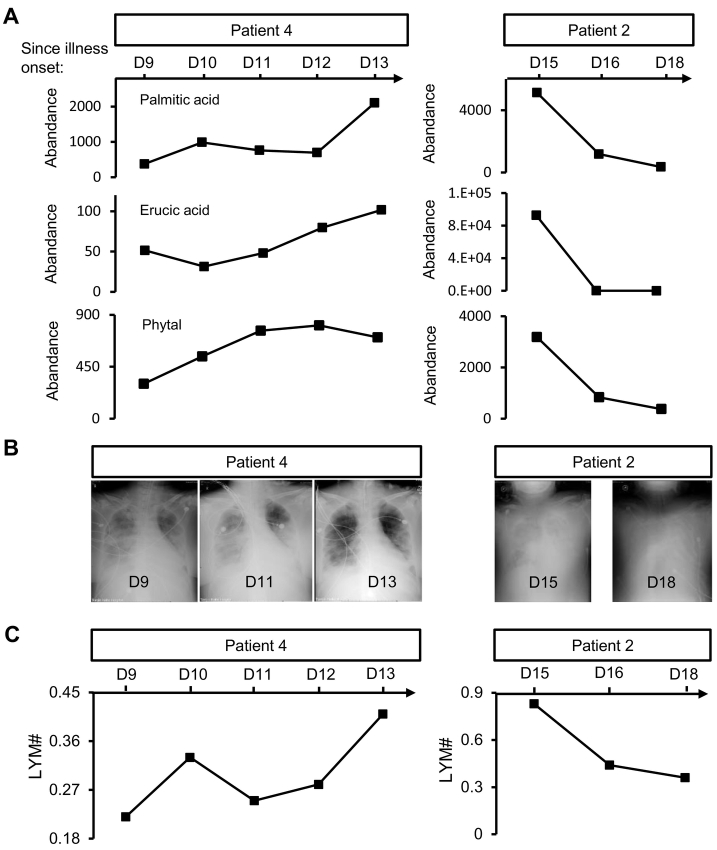
To investigate whether any of these ten identified metabolites is relevant to the prognosis of H7N9-infected patients, we correlated the abundance of serum metabolites with clinical observations over time. We found that the abundance of palmitic acid, erucic acid, and phytal increased in patient 4 from day 9 to day 13 after disease onset (Fig. 6A left column). This period was the time window that reflected the process of resolution of lung inflammation in this patient, as evaluated by chest digital radiography and absolute serum lymphocyte count (Fig. 6B left column, Fig. 6C left column). Absolute lymphocyte count has been reported to decrease after H7N9 infection [11, 29]. Therefore, the increasing trend of absolute lymphocyte count may reflect virus removal, while the decreasing trend of absolute lymphocyte count may reflect viral replication (Fig. 6C). A similar correlation was observed in patient 2, in whom the abundance of palmitic acid, erucic acid, and phytal decreased as lung inflammation became more severe, as evaluated by chest digital radiography and absolute serum lymphocyte count from day 15 to day 18 after disease onset (Fig. 6A right column, Fig. 6B right column, Fig. 6C right column). These data may suggest that the serum levels of palmitic acid, erucic acid, and phytal are negatively correlated to disease severity after H7N9 infection.
Fig. 6.
Potential prognostic metabolites in response to H7N9 infection. (A) Serial chest digital radiography images show the absorption of ground-glass opacity in the lung of patient 4 from day 9 to day 13, while the deterioration of ground-glass opacity is observed in patient 2 from day 15 to day 18 after disease onset. (B) The abundance of five differentially expressed metabolites related to H7N9 infection change over the time course.
4. Discussion
The H7N9 virus is considered one of the most deadly respiratory tract viruses in humans. The clinical, virological, immunological, and epidemiological features of human cases have been reported [8, 16, 24, 52]. To treat such patients, immediate use of antiviral drugs and steroids is very common in the ICU, as is the use of MV or ECMO when necessary [2, 36]. We also observed that epithelial barrier function was severely impaired in the lung biopsy specimen from a patient with laboratory-confirmed H7N9 infection. Metabolomic profiling of serum from four H7N9-infected patients identified ten metabolites that may be related to H7N9 virus infection. Serum abundance of palmitic acid, erucic acid, and phytal may be negatively linked to disease severity. These metabolites may be associated with fatty acid metabolism.
Fatty acid metabolism is closely related to influenza virus infection. Viruses assemble their membrane using the host's unsaturated fatty acids, such as palmitic acid. The spike protein hemagglutinin and the proton-channel M2 of the influenza virus are S-acylated at the cytoplasmic and transmembrane cysteine residues by palmitic acid [31, 49]. Virus particles containing hemagglutinin with removed palmitoylation sites revealed defects in replication and membrane fusion [50, 59]. Although loss of the palmitoylation site in M2 does not affect the production of virus particles, attenuation of virus infectivity was observed when mice were infected with viruses having non-palmitoylated M2 [4, 18]. Therefore, rapid replication of the H7N9 virus consumes a large amount of palmitic acid; this finding is consistent with our observation that the serum level of palmitic acid is significantly reduced during the early onset of disease. In our study, the condition of one patient with a continuous decrease of serum palmitic acid gradually worsened until death. While another patient with an increasing serum level of palmitic acid recovered. Restored serum level of palmitic acid may reflect virus elimination. Therefore, we propose that real-time tracking of the serum level of palmitic acid may predict the prognosis after influenza infection.
Erucic acid, a long-chain monounsaturated fatty acid, is usually found in canola oil, mustard oil, and rapeseed oil. It is digested, absorbed, and metabolized for the most part like other fatty acids in humans [43]. Erucic acid binds to human serum albumin with high affinity; it therefore exists as a complex with albumin [40]. This may explain our observations that both erucic acid and albumin were reduced in the sera of H7N9-infected patients. Erucic acid reduces the synthesis of very-long-chain fatty acids, which are required for efficient viral replication [32]. As observed in the present study, a significant reduction of erucic acid in the early stage of H7N9 infection negatively correlated with the rapid replication of the H7N9 virus. We also found that the serum level of erucic acid positively correlated with the general condition. On the basis of these findings, we propose that the serum level of erucic acid, in addition to that of palmitic acid, may predict the prognosis after H7N9 infection.
The predictive value of phytal, an isoprenoid lipid molecule, is similar to that of palmitic acid and erucic acid. Isoprenoid was shown to inhibit influenza viral infection and growth [44], but the function of phytal in human infections remains unexplored. A previous study showed that sustained high levels of angiotensin II in H7N9-infected patients was strongly correlated with mortality [25]. Besides angiotensin II, C-reactive protein, hypercytokinemia factors, and some clinical parameters such as the PaO2/FiO2 ratio are considered biomarkers for lethality in influenza infections [5, 15, 21]. Comprehensive evaluation of H7N9-infected patients with these serum metabolites, proteins, and clinical parameters should help optimize treatment options for ensuring better outcomes.
Respiratory inflammation is a beneficial process that includes the secretion of antimicrobial products and recruitment of leukocytes to remove inhaled bacteria and viruses. Mucus, which contains mucins such as MUC2, MUC5AC, and MUC5B, is secreted by the tracheobronchial mucosa to cover and protect the epithelial cells from pathogenic microorganisms [30]. Pulmonary surfactant proteins, including SPA, SPB, SPC, and SPD, form a lipid monolayer on the interface of air and liquid and reduce the surface tension of the alveolar epithelium, thereby maintaining the stability of the alveolar structures [53]. Severe inflammation induced by H7N9 infection impairs mucociliary clearance, as observed in the present study by the largely altered expression of both airway mucus and alveolar surfactants unlike in the normal lung. Excess mucus and surfactant proteins not only impair gas conduction and exchange, but also sequester the invaded virus locally, thereby increasing the penetration of the virus into the epithelium [34]. Our preliminary data suggested that tight junctions between the epithelial cells in the lung may be destroyed during H7N9 infection. Epithelial integrity is restored by the endogenous epithelial stem/progenitor cells that reside in different regions of the lung [23, 41]. Upon injuries, these stem/progenitor cells replicate and differentiate to replenish the airway and alveolar cell populations, respectively (Chen et al., [6, 7]; [23]). The influenza virus has been found to interfere with the epithelial stem/progenitor cells in animal studies [33, 39]. These cells may be abrogated during H7N9 infection, so that epithelial damage worsens the ongoing pneumonia, thereby leading to the death of these patients. In addition to traditional treatments, novel therapies, such as stem-cell therapy, could be developed to reduce mortality by enhancing lung regeneration in patients with ARDS [20, 58]. Moreover, small molecules that could boost the regenerative capacity of epithelial stem/progenitor cells should be screened by the drug industry.
Early initiation of antiviral therapy, glucocorticoids, MV, ECMO, CRRT, and other symptomatic and supportive treatments is beneficial in the control of the virus-infection-induced tissue damage. However, two of our patients did not survive such treatments, suggesting that disease progression and treatment outcome may also be determined by other factors. Compared to the two survivors, these two patients had more underlying medical complications, including coronary heart disease, COPD, pulmonary heart disease, or tuberculosis of lymph nodes. The more complicated underlying pathologies make respiratory function more fragile and increase the chance of death, as described previously [16, 27].
The present study has several limitations. First, the number of H7N9-infected patients investigated was low, even though we tried to recruit as many patients as we could. Therefore, the potential metabolites for disease prognosis identified in this study are only suggestive and should be further evaluated in large-scale studies. Second, only one lung biopsy was available for analysis; hence, epithelial damage in the lung caused by H7N9 infection should be further investigated in the future. Third, because of the limited amount of serum collected from human subjects, only UPLC-MS was conducted to analyze the serum metabolites. Other metabolomic tools such as gas chromatography-MS could provide more information about the serum metabolites associated with H7N9 infection. Fourth, the design of control groups was very difficult since various factors could affect the analysis, e.g., predefined criteria for enrolment and exclusion. In addition, the levels of serum metabolites can be affected by a number of host-related factors, including underlying medical conditions (diabetes or other underlying metabolic conditions), mucociliary clearance rate, surfactant and mucin production, and viral/bacterial/fungal co-infections. Therapeutic-intervention-related factors included intubation, MV, ECMO, and variable treatments with antiviral options or different steroids with variable doses and durations. Lastly, the biomarkers identified in the present study, including palmitic acid, erucic acid, and phytal, may not be specific to H7N9 infection, unlike in the case of serum metabolomic analysis of infections with other respiratory viruses including the respiratory syncytial virus and H1N1 virus ([46]; Ferrarini et al., [14]; [10, 47]; Chen et al., [6, 7]). Future re-evaluation will also be needed because all four patients recruited in this study were at the severe end stage of the disease, while those in the control groups were not. It should also be noted that cause and effect cannot be distinguished in this preliminary study because of the lack of data before/after the analyzed time series.
In summary, the present study provided preliminary data suggesting that H7N9 infection may be associated with altered fatty acid metabolism. The potential serum metabolites identified herein may be informative in future evaluations of the prognosis of H7N9-infected patients. Altered structure and secretory properties of the lung epithelium may be associated with the severity of H7N9 virus infections. Novel therapy targeting epithelial regeneration can be a promising tool for the effective control of H7N9 infection.
Acknowledgments
Acknowledgements
Not applicable.
Declaration of Interest
The authors indicate no potential conflicts of interest.
Author Contributions
HC conceived, designed, and supervised the study. XS, LS, SF, LL, HY, QWang, XW, ZH, XL, YL, QZ, KL, CC, JW, and ZQ collected data. XS, LS, SF, QWu, and HC analyzed the data. LS, SF, and HC wrote the drafts of the manuscript. XS, LS, SF, LL, HY, QWu, and HC interpreted the findings. XW, ZH, XL, YL, QZ, KL, CC, JW, and ZQ commented on and revised the drafts of the manuscript. All authors read and approved the final report.
Funding
This study was supported by the National Natural Science Foundation of China (31471121, 81773394, and 81728001), the Natural Science Foundation of Tianjin (17JCYBJC24700), and Key Projects of Health and Family Planning Commission of Tianjin (16KG163 and 16KG164).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.06.019.
Contributor Information
Qi Wu, Email: wq572004@163.com.
Huaiyong Chen, Email: huaiyong.chen@foxmail.com.
Appendix A. Supplementary Data
Supplementary material
References
- 1.Bertran K., Lee D.H., Pantin-Jackwood M.J., Spackman E., Balzli C., Suarez D.L. Pathobiology of Clade 2.3.4.4 H5Nx high-pathogenicity avian influenza virus infections in minor gallinaceous poultry supports early backyard flock introductions in the Western United States in 2014-2015. J Virol. 2017:91. doi: 10.1128/JVI.00960-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao B., Gao H., Zhou B., Deng X., Hu C., Deng C. Adjuvant corticosteroid treatment in adults with influenza A (H7N9) viral pneumonia. Crit Care Med. 2016;44:e318–e328. doi: 10.1097/CCM.0000000000001616. [DOI] [PubMed] [Google Scholar]
- 3.Cao H.F., Liang Z.H., Feng Y., Zhang Z.N., Xu J., He H. A confirmed severe case of human infection with avian-origin influenza H7N9: a case report. Exp Ther Med. 2015;9:693–696. doi: 10.3892/etm.2014.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castrucci M.R., Hughes M., Calzoletti L., Donatelli I., Wells K., Takada A. The cysteine residues of the M2 protein are not required for influenza A virus replication. Virology. 1997;238:128–134. doi: 10.1006/viro.1997.8809. [DOI] [PubMed] [Google Scholar]
- 5.Charles P.G. Early diagnosis of lower respiratory tract infections (point-of-care tests) Curr Opin Pulm Med. 2008;14:176–182. doi: 10.1097/MCP.0b013e3282f7642f. [DOI] [PubMed] [Google Scholar]
- 6.Chen H., Matsumoto K., Brockway B.L., Rackley C.R., Liang J., Lee J.H. Airway epithelial progenitors are region specific and show differential responses to bleomycin-induced lung injury. Stem Cells. 2012;30:1948–1960. doi: 10.1002/stem.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen R., Mias G.I., Li-Pook-Than J., Jiang L., Lam H.Y., Chen R. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y., Liang W., Yang S., Wu N., Gao H., Sheng J. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013;381:1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui L., Fang J., Ooi E.E., Lee Y.H. Serial metabolome changes in a prospective cohort of subjects with influenza viral infection and comparison with dengue fever. J Proteome Res. 2017;16:2614–2622. doi: 10.1021/acs.jproteome.7b00173. [DOI] [PubMed] [Google Scholar]
- 10.Cui L., Zheng D., Lee Y.H., Chan T.K., Kumar Y., Ho W.E. Metabolomics investigation reveals metabolite mediators associated with acute lung injury and repair in a murine model of influenza pneumonia. Sci Rep. 2016;6:26076. doi: 10.1038/srep26076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diao H., Cui G., Wei Y., Chen J., Zuo J., Cao H. Severe H7N9 infection is associated with decreased antigen-presenting capacity of CD14+ cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan J., Cui D., Lau S., Xie G., Guo X., Zheng S. Detection of a novel avian influenza A (H7N9) virus in humans by multiplex one-step real-time RT-PCR assay. BMC Infect Dis. 2014;14:541. doi: 10.1186/1471-2334-14-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feizi N., Mehrbod P., Romani B., Soleimanjahi H., Bamdad T., Feizi A. Autophagy induction regulates influenza virus replication in a time-dependent manner. J Med Microbiol. 2017;66:536–541. doi: 10.1099/jmm.0.000455. [DOI] [PubMed] [Google Scholar]
- 14.Ferrarini A., Righetti L., Martínez M.P., Fernández-López M., Mastrangelo A., Horcajada J.P. Discriminant biomarkers of acute respiratory distress syndrome associated to H1N1 influenza identified by metabolomics HPLC-QTOF-MS/MS platform. Electrophoresis. 2017;38:2341–2348. doi: 10.1002/elps.201700112. [DOI] [PubMed] [Google Scholar]
- 15.Fine M.J., Auble T.E., Yealy D.M., Hanusa B.H., Weissfeld L.A., Singer D.E. A prediction rule to identify low-risk patients with community-acquired pneumonia. N Engl J Med. 1997;336:243–250. doi: 10.1056/NEJM199701233360402. [DOI] [PubMed] [Google Scholar]
- 16.Gao H.N., Lu H.Z., Cao B., Du B., Shang H., Gan J.H. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013;368:2277–2285. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
- 17.Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 18.Grantham M.L., Wu W.H., Lalime E.N., Lorenzo M.E., Klein S.L., Pekosz A. Palmitoylation of the influenza A virus M2 protein is not required for virus replication in vitro but contributes to virus virulence. J Virol. 2009;83:8655–8661. doi: 10.1128/JVI.01129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gubareva L.V., Sleeman K., Guo Z., Yang H., Hodges E., Davis C.T. Drug susceptibility evaluation of an influenza A(H7N9) virus by analyzing recombinant neuraminidase proteins. J Infect Dis. 2017;216:S566–S574. doi: 10.1093/infdis/jiw625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guillamat-Prats R., Camprubi-Rimblas M., Bringue J., Tantinya N., Artigas A. Cell therapy for the treatment of sepsis and acute respiratory distress syndrome. Ann Transl Med. 2017;5:446. doi: 10.21037/atm.2017.08.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo J., Huang F., Liu J., Chen Y., Wang W., Cao B. The serum profile of Hypercytokinemia factors identified in H7N9-infected patients can predict fatal outcomes. Sci Rep. 2015;5:10942. doi: 10.1038/srep10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henry Dunand C.J., Leon P.E., Huang M., Choi A., Chromikova V., Ho I.Y. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe. 2016;19:800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogan B.L., Barkauskas C.E., Chapman H.A., Epstein J.A., Jain R., Hsia C.C. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15:123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y., Lu S., Song Z., Wang W., Hao P., Li J. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet. 2013;381:2273–2279. doi: 10.1016/S0140-6736(13)61125-3. [DOI] [PubMed] [Google Scholar]
- 25.Huang F., Guo J., Zou Z., Liu J., Cao B., Zhang S. Angiotensin II plasma levels are linked to disease severity and predict fatal outcomes in H7N9-infected patients. Nat Commun. 2014;5:3595. doi: 10.1038/ncomms4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hui D.S.C., Lee N., Chan P.K.S. A clinical approach to the threat of emerging influenza viruses in the Asia-Pacific region. Respirology. 2017;22:1300–1312. doi: 10.1111/resp.13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Husain M. Avian influenza A (H7N9) virus infection in humans: epidemiology, evolution, and pathogenesis. Infect Genet Evol. 2014;28:304–312. doi: 10.1016/j.meegid.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 28.Imai M., Watanabe T., Kiso M., Nakajima N., Yamayoshi S., Iwatsuki-Horimoto K. A highly pathogenic avian H7N9 influenza virus isolated from a human is lethal in some ferrets infected via respiratory droplets. Cell Host Microbe. 2017;22:615–626. doi: 10.1016/j.chom.2017.09.008. [e618] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji H., Gu Q., Chen L.L., Xu K., Ling X., Bao C.J. Epidemiological and clinical characteristics and risk factors for death of patients with avian influenza A H7N9 virus infection from Jiangsu Province, Eastern China. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knowles M.R., Boucher R.C. Mucus clearance as a primary innate defense mechanism for mammalian airways. J Clin Invest. 2002;109:571–577. doi: 10.1172/JCI15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kordyukova L.V., Ksenofontov A.L., Serebryakova M.V., Ovchinnikova T.V., Fedorova N.V., Ivanova V.T. Influenza A hemagglutinin C-terminal anchoring peptide: identification and mass spectrometric study. Protein Pept Lett. 2004;11:385–391. doi: 10.2174/0929866043406850. [DOI] [PubMed] [Google Scholar]
- 32.Koyuncu E., Purdy J.G., Rabinowitz J.D., Shenk T. Saturated very long chain fatty acids are required for the production of infectious human cytomegalovirus progeny. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar P.A., Hu Y., Yamamoto Y., Hoe N.B., Wei T.S., Mu D. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011;147:525–538. doi: 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leiva-Juarez M.M., Kolls J.K., Evans S.E. Lung epithelial cells: therapeutically inducible effectors of antimicrobial defense. Mucosal Immunol. 2018;11:21–34. doi: 10.1038/mi.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma W., Huang H., Chen J., Xu K., Dai Q., Yu H. Predictors for fatal human infections with avian H7N9 influenza, evidence from four epidemic waves in Jiangsu Province, eastern China, 2013-2016. Influenza Other Respi Viruses. 2017;11:418–424. doi: 10.1111/irv.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nie Q., Zhang D.Y., Wu W.J., Huang C.L., Ni Z.Y. Extracorporeal membrane oxygenation for avian influenza A (H7N9) patient with acute respiratory distress syndrome: a case report and short literature review. BMC Pulm Med. 2017;17:38. doi: 10.1186/s12890-017-0381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan H., Zhang X., Hu J., Chen J., Pan Q., Teng Z. A case report of avian influenza H7N9 killing a young doctor in Shanghai, China. BMC Infect Dis. 2015;15:237. doi: 10.1186/s12879-015-0970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian L., Zheng J., Xu H., Shi L., Li L. Extracorporeal membrane oxygenation treatment of a H7N9-caused respiratory failure patient with mechanical valves replacement history: a case report. Med (Baltimore) 2016;95 doi: 10.1097/MD.0000000000005052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quantius J., Schmoldt C., Vazquez-Armendariz A.I., Becker C., El Agha E., Wilhelm J. Influenza virus infects epithelial stem/progenitor cells of the distal lung: impact on Fgfr2b-driven epithelial repair. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rabbani G., Baig M.H., Jan A.T., Ju Lee E., Khan M.V., Zaman M. Binding of erucic acid with human serum albumin using a spectroscopic and molecular docking study. Int J Biol Macromol. 2017;105:1572–1580. doi: 10.1016/j.ijbiomac.2017.04.051. [DOI] [PubMed] [Google Scholar]
- 41.Rackley C.R., Stripp B.R. Building and maintaining the epithelium of the lung. J Clin Invest. 2012;122:2724–2730. doi: 10.1172/JCI60519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shenoy M.K., Iwai S., Lin D.L., Worodria W., Ayakaka I., Byanyima P. Immune response and mortality risk relate to distinct lung microbiomes in patients with HIV and pneumonia. Am J Respir Crit Care Med. 2017;195:104–114. doi: 10.1164/rccm.201603-0523OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin D.S., Perlman S., Rosove M.H. Romiplostim mitigates dose-limiting thrombocytopenia of erucic acid for adrenomyeloneuropathy. Br J Haematol. 2015;171:879–881. doi: 10.1111/bjh.13440. [DOI] [PubMed] [Google Scholar]
- 44.Shoji M., Arakaki Y., Esumi T., Kohnomi S., Yamamoto C., Suzuki Y. Bakuchiol is a phenolic isoprenoid with novel enantiomer-selective anti-influenza A virus activity involving Nrf2 activation. J Biol Chem. 2015;290:28001–28017. doi: 10.1074/jbc.M115.669465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart C.J., Skeath T., Nelson A., Fernstad S.J., Marrs E.C., Perry J.D. Preterm gut microbiota and metabolome following discharge from intensive care. Sci Rep. 2015;5:17141. doi: 10.1038/srep17141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart C.J., Hasegawa K., Wong M.C., Ajami N.J., Petrosino J.F., Piedra P.A. Respiratory syncytial virus and rhinovirus bronchiolitis are associated with distinct metabolic pathways. J Infect Dis. 2018;217:1160–1169. doi: 10.1093/infdis/jix680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tisoncik-Go J., Gasper D.J., Kyle J.E., Eisfeld A.J., Selinger C., Hatta M. Integrated omics analysis of pathogenic host responses during pandemic H1N1 influenza virus infection: the crucial role of lipid metabolism. Cell Host Microbe. 2016;19:254–266. doi: 10.1016/j.chom.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Riel D., Leijten L.M.E., de Graaf M., Siegers J.Y., Short K.R., Spronken M.I.J. Novel avian-origin influenza A (H7N9) virus attaches to epithelium in both upper and lower respiratory tract of humans. Am J Pathol. 2013;183:1137–1143. doi: 10.1016/j.ajpath.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veit M., Serebryakova M.V., Kordyukova L.V. Palmitoylation of influenza virus proteins. Biochem Soc Trans. 2013;41:50–55. doi: 10.1042/BST20120210. [DOI] [PubMed] [Google Scholar]
- 50.Wagner R., Herwig A., Azzouz N., Klenk H.D. Acylation-mediated membrane anchoring of avian influenza virus hemagglutinin is essential for fusion pore formation and virus infectivity. J Virol. 2005;79:6449–6458. doi: 10.1128/JVI.79.10.6449-6458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H., Xiao X., Lu J., Chen Z., Li K., Liu H. Factors associated with clinical outcome in 25 patients with avian influenza A (H7N9) infection in Guangzhou, China. BMC Infect Dis. 2016;16:534. doi: 10.1186/s12879-016-1840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X., Jiang H., Wu P., Uyeki T.M., Feng L., Lai S. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013-17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis. 2017;17:822–832. doi: 10.1016/S1473-3099(17)30323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitsett J.A., Wert S.E., Weaver T.E. Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annu Rev Med. 2010;61:105–119. doi: 10.1146/annurev.med.60.041807.123500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L., Zhu W., Li X., Chen M., Wu J., Yu P. Genesis and spread of newly emerged highly pathogenic H7N9 avian viruses in mainland China. J Virol. 2017;91 doi: 10.1128/JVI.01277-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeganeh B., Ghavami S., Rahim M.N., Klonisch T., Halayko A.J., Coombs K.M. Autophagy activation is required for influenza A virus-induced apoptosis and replication. Biochim Biophys Acta. 2018;1865:364–378. doi: 10.1016/j.bbamcr.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 56.Yu L., Wang Z., Chen Y., Ding W., Jia H., Chan J.F. Clinical, virological, and histopathological manifestations of fatal human infections by avian influenza A(H7N9) virus. Clin Infect Dis. 2013;57:1449–1457. doi: 10.1093/cid/cit541. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y., Gao H., Liang W., Tang L., Yang Y., Wu X. Efficacy of oseltamivir-peramivir combination therapy compared to oseltamivir monotherapy for influenza A (H7N9) infection: a retrospective study. BMC Infect Dis. 2016;16:76. doi: 10.1186/s12879-016-1383-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu Y., Chen X., Yang X., Ji J., Hashash A. Stem cells in lung repair and regeneration: current applications and future promise. J Cell Physiol. 2017 Dec 22 doi: 10.1002/jcp.26414. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 59.Zurcher T., Luo G., Palese P. Mutations at palmitylation sites of the influenza virus hemagglutinin affect virus formation. J Virol. 1994;68:5748–5754. doi: 10.1128/jvi.68.9.5748-5754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material