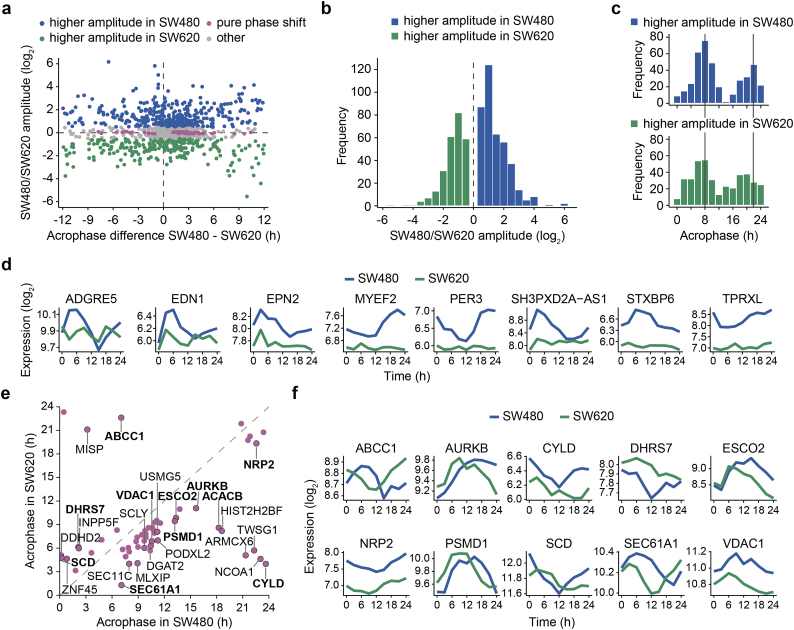
Fig. 2.
Circadian transcripts exhibit differential rhythmicity in the CRC progression model.
(a) Changes in rhythmicity (amplitudes and/or acrophases) from the primary tumor cell line SW480 to the metastatic cell line SW620 of the transcripts that showed 24-h rhythms in either one of the cell lines were estimated using the robust DODR method. The transcripts with BH adjusted p < 0·05 were further divided in transcripts with a higher amplitude in SW480 cells (blue, log2 amplitude change >0·5), a higher amplitude in SW620 cells (green, log2 amplitude change < −0·5) and transcripts with a pure phase shift (pink, FC amplitude >1·15 in both cell lines, absolute log2 amplitude change <0·1 and phase shift >1 h). (b) log2 amplitude changes from SW480 to SW620 cells for transcripts with DODR adjusted p < 0·05 and absolute log2 amplitude change >0·5. (c) Acrophases for transcripts with higher amplitudes in SW480 cells (upper panel, blue) and SW620 cells (lower panel, green). (d) Time-course for the eight transcripts with the largest loss of oscillations in SW620 cells (absolute log2 amplitude change >4, FC amplitude >1·25 in SW480 cells). (e) Acrophases in SW480 and SW620 cells for transcripts with pure phase shifts. Transcripts with absolute phase shifts >3 h (black border) are labeled with the gene name. Selected transcripts represented in (f) are written in bold. (f) Time-course for selected phase-shifted transcripts.