Fig. 3.
Circadian pathway analysis identifies temporally coordinated expression of spliceosome-related genes in CRC cell lines.
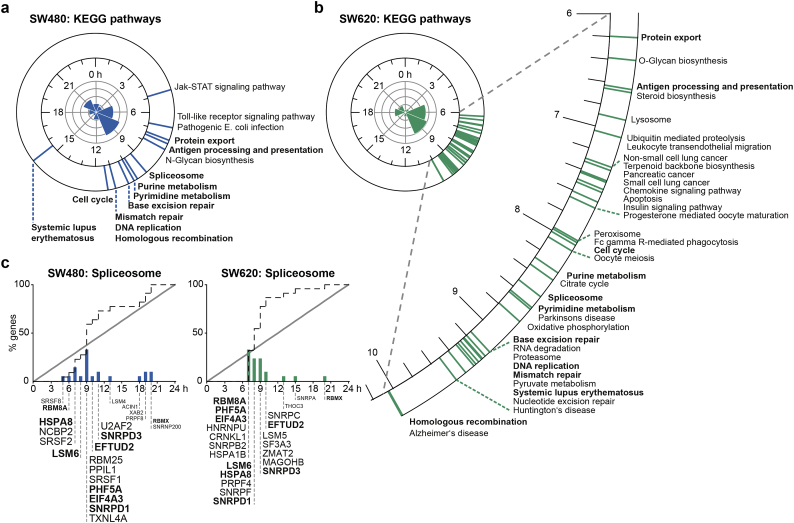
Summary of significantly phase-clustered (q < 0·01) circadian pathways in SW480 (a) and SW620 cells (b). The circular axis (blue lines: SW480; green lines: SW620) represents the average phase of all circadian genes in a pathway from the KEGG database. Pathways that are temporally coordinated in both cell lines are written in bold. The circular histograms show the 3-h acrophase bins of the 24-h cycling transcripts. (c) The spliceosome is among the phase-clustered pathways that demonstrated circadian coordination in both SW480 (left panel) and SW620 cells (right panel) cells. The dotted line depicts the empirical cumulative distribution of phases belonging to genes that are part of the spliceosome pathway. The solid line depicts the uniform distribution. The height of the bars corresponds to the percentage of circadian genes in the spliceosome pathway that peak at a certain time after synchronization. Individual gene names are shown with a larger font size corresponding to a greater contribution of the gene to the overall phase clustering of the pathway. Gene acrophases were rounded to the full hour.