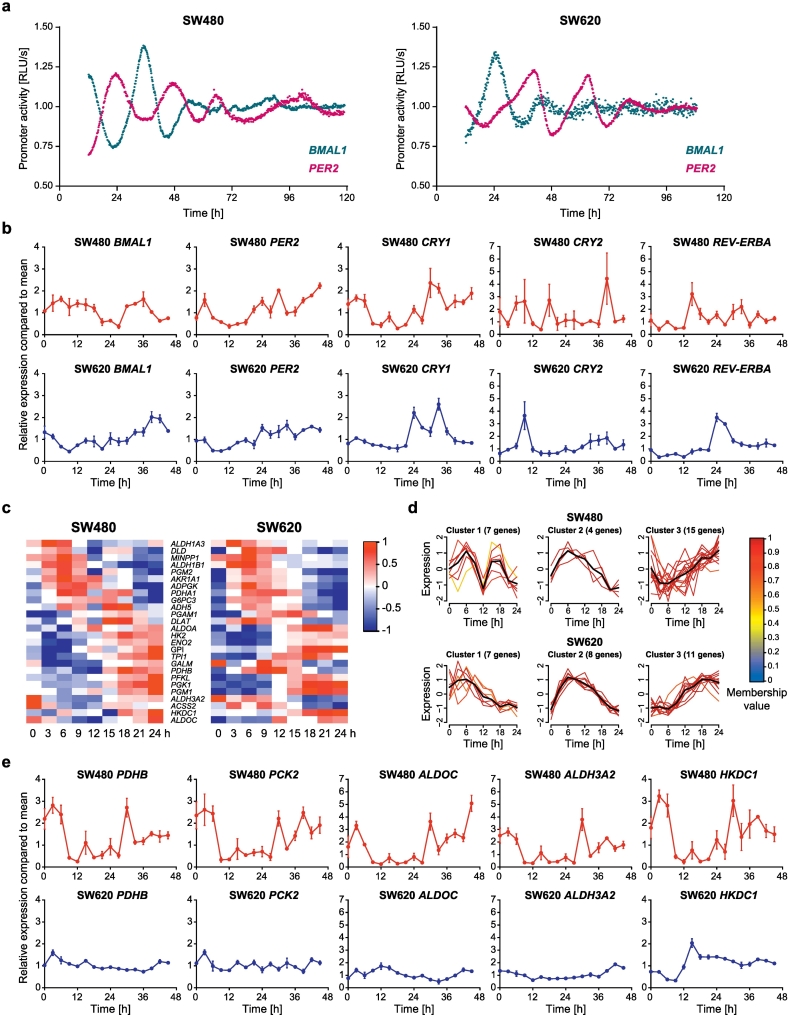
Fig. 1.
SW480 and SW620 cells serve as a model for tumor progression with changes in the core-clock and metabolic output pathways.
(a) To investigate circadian clock activity, SW480 (left panel) and SW620 (right panel) cells were lentivirally transduced with a BMAL1-promoter (blue) or PER2-promoter (pink) driven luciferase construct. Bioluminescence was measured for five consecutive days. Shown is one representative replicate for each condition. (b) 45 h time-course RT-qPCR measurements of selected core-clock genes (BMAL1, CRY1, CRY2, PER2 and REV-ERBα) in SW480 (dark red) and SW620 (dark blue) cells. Data are expressed as mean ± SEM, n = 3. (c) Phase-ordered, median-normalized heatmap of glycolytic genes oscillating with a 24 h period in at least one of the two tested cell lines. (d) Clustering of metabolic genes based on their time-dependent expression profile. The colors of the gene expression patterns indicate gradual membership values, reflecting the strength of a gene's association with its respective cluster (red: high membership value, blue: low membership value). The black lines indicate the cluster centres. (e) 45 h time-course RT-qPCR measurements of selected core-clock genes (ALDH3A2, ALDOC, HKDC1, PCK2 and PDHB) in SW480 (dark red) and SW620 (dark blue) cells. Data are expressed as mean ± SEM, n = 3. See also Supplementary Fig 1 and 2 and Supplementary Tables 1 and 2.