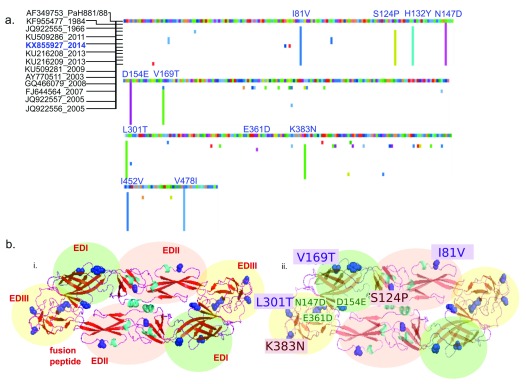
Figure 3. Shared amino acid substitutions in the envelope protein of Indian DENV3 strains differ from PaH881/8.
( a) Multiple sequence alignment of region coding for the envelope (E) protein of dengue virus 3 from India were aligned to gi|13310784|gb|AF349753.1| DENV3 strain PaH881/88 polyprotein precursor, translated E genes. Suffix represent the year of sampling. Predicted amino acid changes compared to PaH881/88 are shown in colour. Position of substitutions present in the sequenced KX855927 strain are shown in blue. ( b) i) Cartoon structure of E protein KX855927 (2014)- dimer, homology modeled in SWISS-PROT with the domains shaded green (EDI), pink (EDII) and yellow (EDIII), labeled in red. ii) Cartoon structure of E protein KX855927 (2014)- dimer, homology modeled in SWISS-PROT showing the amino acid substitutions in KX855927 (2014) compared to the PaH881/8 in one of the dimers. In both cartoons, predicted substitutions are shown in blue (side-chains colored). Amino acid substitutions labelled in violet (violet box) are positions known to influence mouse monoclonal antibody binding. Positions in red (red box) are among 32 positions in the E protein predicted to be important for antigenicity.