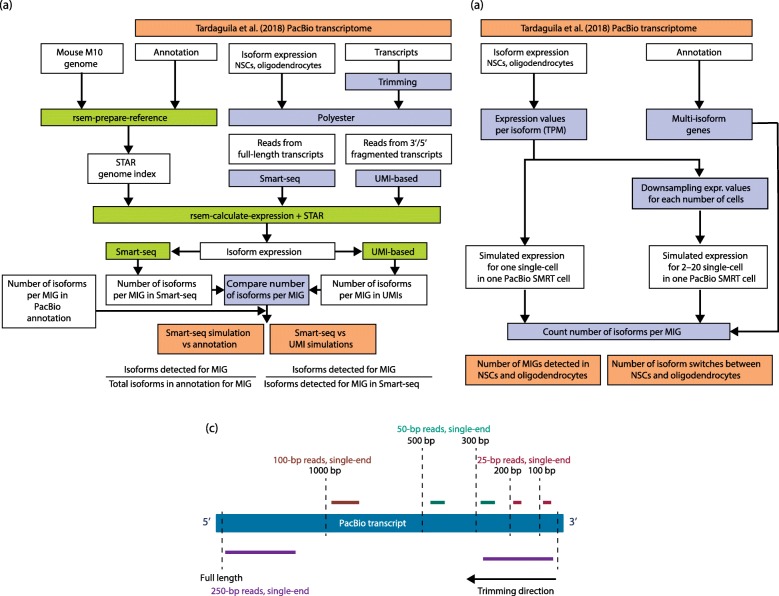
Fig. 4.
Simulation of short- and long-read workflows and the modelling of a UMI-based library preparation strategy. a Short-read simulation workflow. Transcript sequences from the Tardaguila et al. 2018 neural transcriptome [66] were trimmed, and reads simulated from fragments to recreate UMI library preparation limitations in transcript covered length. Full-length reads were also simulated. Reads were aligned to the mouse genome using STAR and isoform expression quantified using RSEM. For UMI simulations, the number of isoforms resolved using Smart-seq reads was used as the 100% reference to calculate the percentage of resolution of MIG. For the Smart-seq simulation, the annotated number of isoforms per gene (in Tardaguila et al. [66]) was used as the 100% reference. b Long-read simulation workflow. The Illumina quantification of isoform expression available in Tardaguila et al. [66] was scaled to one million reads (TPM) to recreate a Sequel run of one million long reads, where a single cell is sequenced. Values were downsampled to simulate scenarios where an increasing number of cells (2, 6, 10, 16, 20) are sequenced together in a similar run. The number of reads per cell is therefore gradually decreasing. The number of MIGs in the Tardaguila et al. annotation was compared with the number of MIGs detected in the simulated scenarios. Then, the number of isoform switches detected in the Tardaguila et al. data was compared. c Short-read length simulated for each simulation scenario (represented for 3′ UMIs only). PacBio transcript sequences in the Tardaguila et al. dataset [66] were trimmed as described. To ensure that coverage was even when capturing growing lengths of the transcripts in simulated UMI-based protocols, the length of the simulated reads was increased for longer fragments (100 and 200 bp—25 bp reads, 300 and 500 bp—50 bp reads, 1000 bp—100 bp reads, full length—250 bp reads, paired-end). MIG multi-isoform gene, NSC neural stem cell, RSEM RNA-seq by expectation maximization, TPM transcripts per million, UMI unique molecular identifier