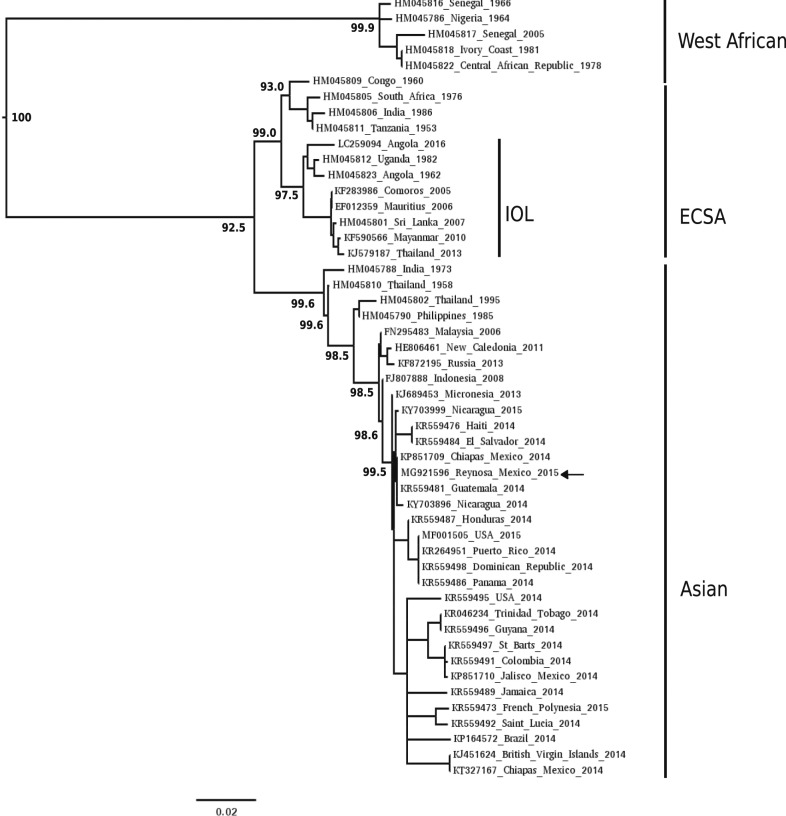
Figure 2.
Phylogenetic analysis of the complete genome sequences of CH-R-1950 and other select chikungunya virus (CHIKV) isolates. Nucleotide sequences were aligned using the program multiple alignment using fast Fourier transform. A Markov Chain Monte Carlo–based Bayesian analysis was performed by MrBayes using the general time reversible plus invariant sites plus gamma distribution 4 model.20 Four million iterations were used to reach convergence. Select posterior probabilities are denoted as percentages under nodes. Branch lengths are proportional to the number of nucleotide differences. The Asian, East/Central/South African (ECSA), Indian Ocean, and West African lineages of CHIKV are denoted.