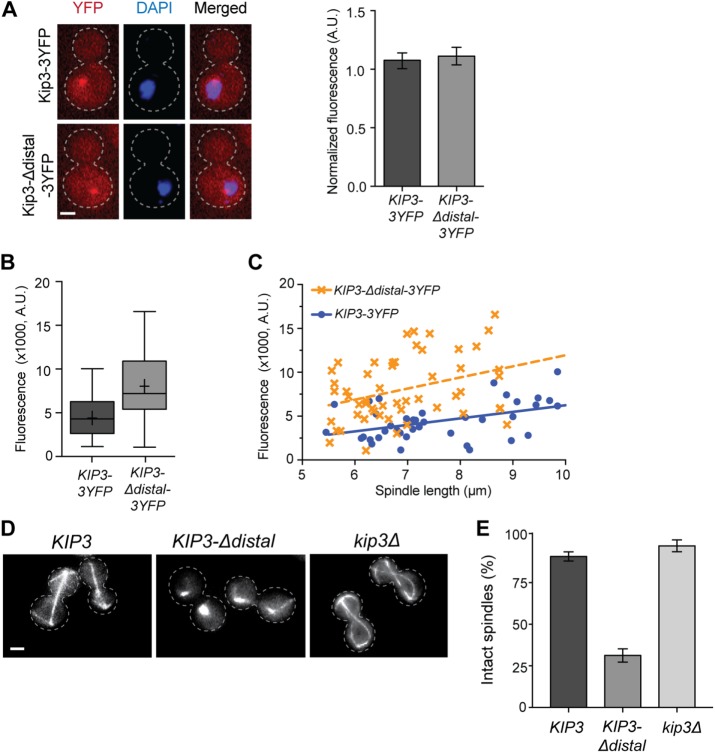
FIGURE 7:
The distal tail of Kip3 temporally regulates spindle disassembly during anaphase. (A) Removal of the distal tail does not alter Kip3 distribution between cytoplasm and nucleus. Left, Single focal plane fluorescence images of cells expressing 3YFP-tagged Kip3 or Kip3-Δdistal (red) following microtubule depolymerization by 15-min nocodazole treatment. Cell outline is indicated by a dotted line; DNA is stained with DAPI (blue); the bright YFP foci is Kip3/Kip3-Δdistal associated with the spindle pole body and/or kinetochores. Right, graph depicting the nuclear-to-cytoplasmic ratio of fluorescence signal for Kip3-3YFP and Kip3-Δdistal-3YFP. Mean ± SD. (B) Quantification of Kip3-3YFP or Kip3-Δdistal-3YFP localized to the spindle midzone region. Center line is median, boxes represent the 25th to 75th percentiles, and whiskers encompass from minimum to maximum values; (+) represents mean values; p = 0.002. (C) Kip3-3YFP or Kip3-Δdistal-3YFP localization to the spindle midzone region relative to spindle length. Both Kip3 and Kip3-Δdistal are positively correlated with spindle length. Dashed lines represent best fit linear regression, and each symbol represents a separate spindle. R2 = 0.20 and 0.12 for KIP3 and KIP3-Δdistal; p < 0.0001 for the difference in elevation (fluorescence intensity) between the lines. The difference between slopes is not statistically significant. (D) Images of microtubules (GFP-Tub1) in KIP3, KIP3-Δdistal, and kip3Δ cells during anaphase arrest (cdc15-2, 2 h at 37°C). Note that spindles are disassembled in KIP3-Δdistal cells. Cell outline indicated by dotted line. (E) Quantification of spindle stability during anaphase arrest in cells of the indicated genotype. Mean ± SD; p < 0.0001 for KIP3-Δdistal vs. either cell type. (A, D) Bars, 2 μm; (A) n = 62 for both; (B, C) n = 39 for Kip3-3YFP and 53 for Kip3-Δdistal-3YFP; (E) graph shows three experiments where n = 313, 281, 175 for KIP3; 259, 266, 275 for KIP3-Δdistal; and 196, 95, 148 for kip3Δ.