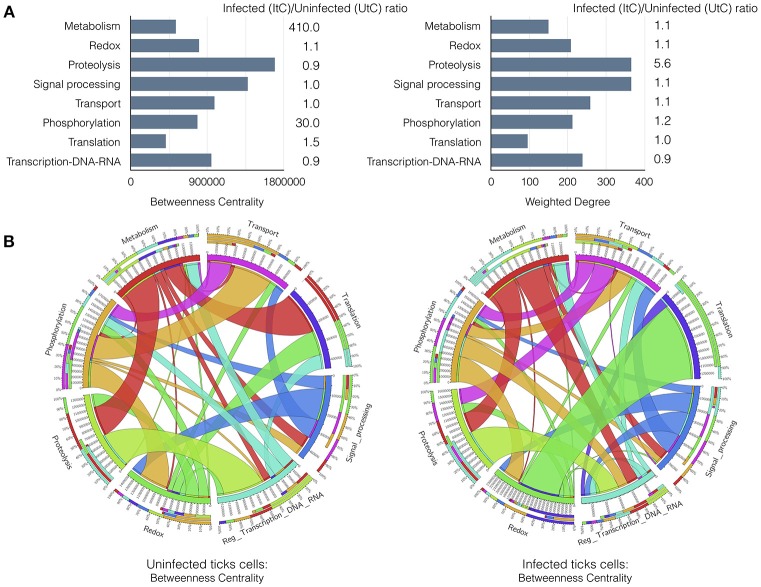
Figure 2.
Results of the graph analysis in A. phagocytophilum infected and uninfected tick vector cells. (A) Variations in Betweenness Centrality (BNC) and Weighted Degree (WD) values between UtC and ItC. Bars represent BNC and WD values for UtC. The fold change between ItC and UtC is included near each bar. (B) Comparison of the WD between UtC and ItC for the proteins of each biological pathway and their connections to other biological pathways. A circle layout displaying the relationships between biological pathways and proteins linking them in both UtC and ItC is shown. Each sector of the circle shows the names of the eight most important biological pathways detected by a clustering algorithm. The first sector below the name shows the percentage of proteins of that biological pathway present in other biological pathways. The second sector indicates the percentage of proteins belonging to other biological pathways that also affect the biological pathway of reference. The third sector displays the percentage of proteins that belong to that biological pathway and are also involved in other biological pathways. The fourth sector displays the actual value of the parameter displayed in the figure. Colors for these three sectors indicate the set of proteins from/to different biological pathways that are shared. The white band above the fourth sector indicates the proportion of “in-coming proteins” while the rest of the sector is used to display the proportion of “out-going proteins”. The relative size of each band is proportional to the WD of the proteins shared among the biological pathways. Note: The Supplementary Figure 3 displays an example with annotations and labels to interpret the circle and band layouts.