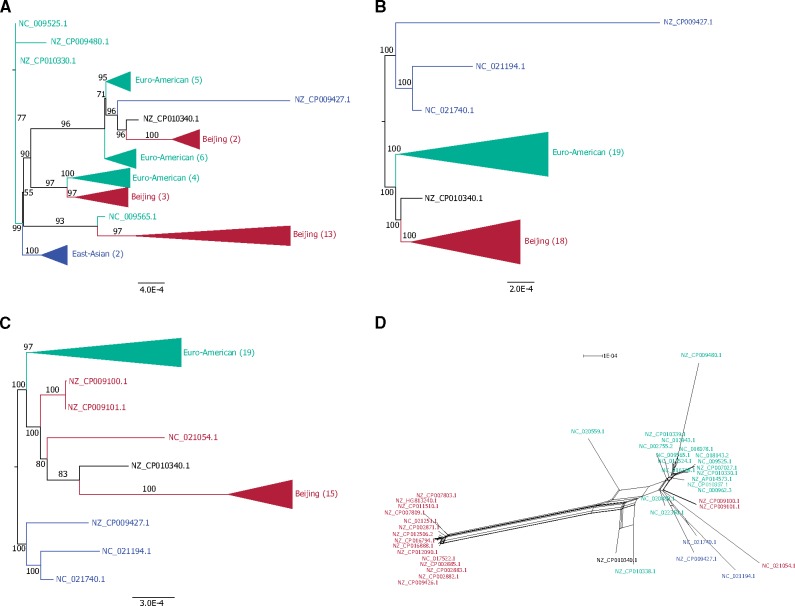
Fig. 3.
—Examples of incongruent phylogenies. (A) Phylogeny of universal genomic region 35 inferred from the complete alignment (1,164,225 nt, 1.2% variable sites, 0.92% parsimony informative sites, HoT score: 50.41%). (B) Phylogeny of universal genomic region 35 estimated from the alignment with unreliable positions removed (586,857 nt, 0.71% variable sites, 0.43% parsimony informative sites). See also supplementary figure S3, Supplementary Material online. (C) Phylogeny of universal genomic region 40 inferred from the complete alignment (611,340 nt, 0.68% variable sites, 0.46% parsimony informative sites, HoT score: 98.28%). (D) Splits network of universal genomic region 40 with unreliable positions removed (600,805 nt, 0.62% variable sites, 0.41% parsimony informative sites). See also supplementary figure S5, Supplementary Material online. Five problematic strains are identified: NZ_CP010340.1 and NZ_CP010338.1 were removed from RefSeq, and NZ_CP009100.1, NZ_CP009101.1, and NC_021054.1 from lineage 2 are H37Rv-guided assemblies.