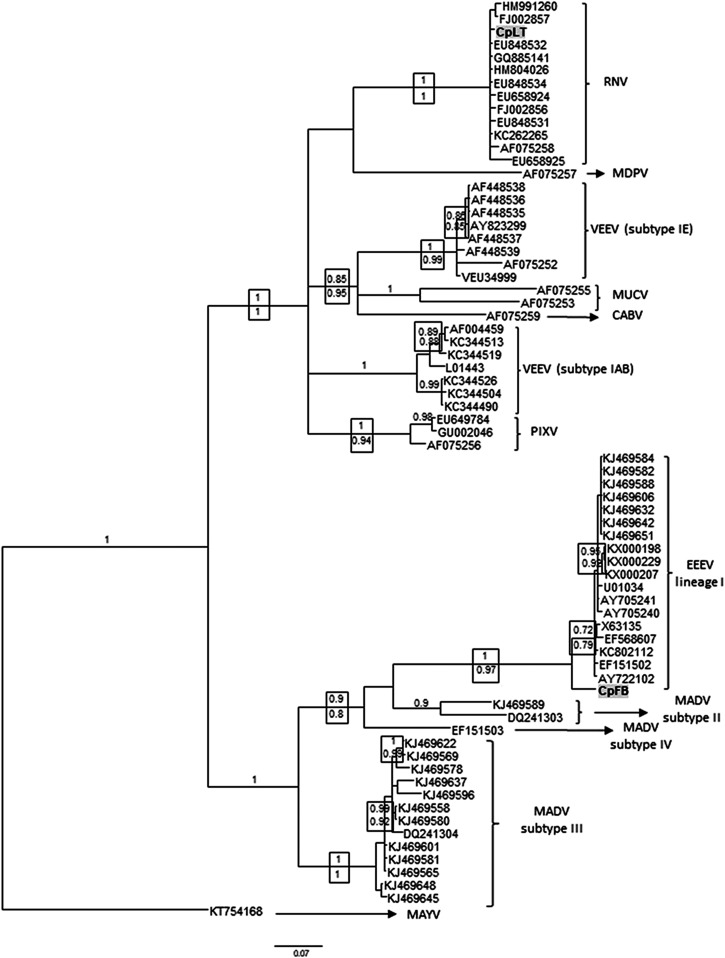
Figure 2.
Bayesian phylogenetic analysis of alphavirus detected in Uruguay. Partial nsP4 genome sequences from mosquito samples were analyzed in comparison with sequences of the VEEV and EEEV complex. The analysis was conducted under the general time reversible + gamma + proportion invariant (GTR + Γ + I) model of nucleotide substitution. Two runs of four chains each (one cold and three heated, temperature 0.20) were run for three million generations; trees were sampled every 100 generations. Convergence was assessed by using the average standard deviation in partition frequency values across independent analyses with a threshold value of 0.01; burn-in was set to 25%. Supports above nodes are the posterior probabilities and below are depicted the approximate likelihood ratio test supports from maximum likelihood analysis. GenBank accession numbers: CpFB: MG009260 and CpLT: MG009261. CABV = Cabassou virus; EEEV = eastern equine encephalitis virus; MADV = Madariaga virus; MAYV = Mayaro virus; MDPV = Mosso das Pedras virus; MUCV = Mucambo virus; PIXV = Pixuna virus; RNV = Rio Negro virus; and VEEV = Venezuelan equine encephalitis virus.