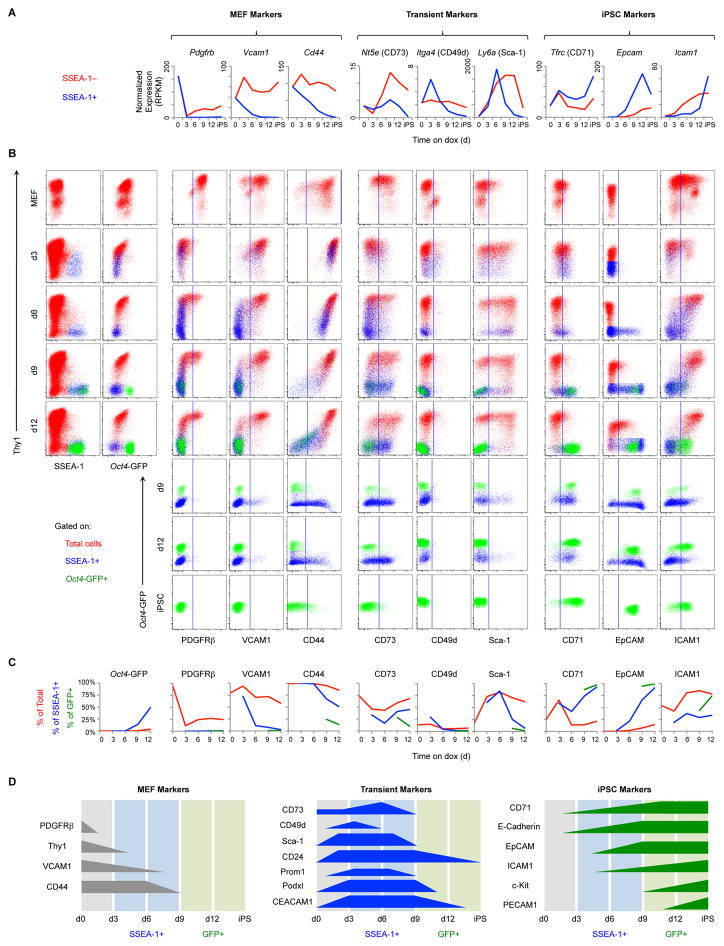
Figure 2. Identification and Characterization of Reprogramming Markers.
(A) Transcriptional profiles of candidate cell-surface markers. SSEA-1− (red) and SSEA-1+ intermediates (blue) derived from het/het MEFs were analyzed by RNA-seq. Results are shown as the mean of 3 replicates.
(B) Flow analysis of het/het MEF reprogramming. Representative dot plots are gated on total live cells (red), SSEA-1+ cells (blue), or Oct4-GFP+ cells (green). Vertical lines indicate the cutoff between negative and positive staining based on an isotype-matched negative control.
(C) Quantification of the plots in Figure 2B. Lines show the percentage of total live cells (red), SSEA-1+ cells (blue), and Oct4-GFP+ cells (green) positive for the indicated markers.
(D) Schematic of reprogramming marker expression based on the results above and Figure S2.