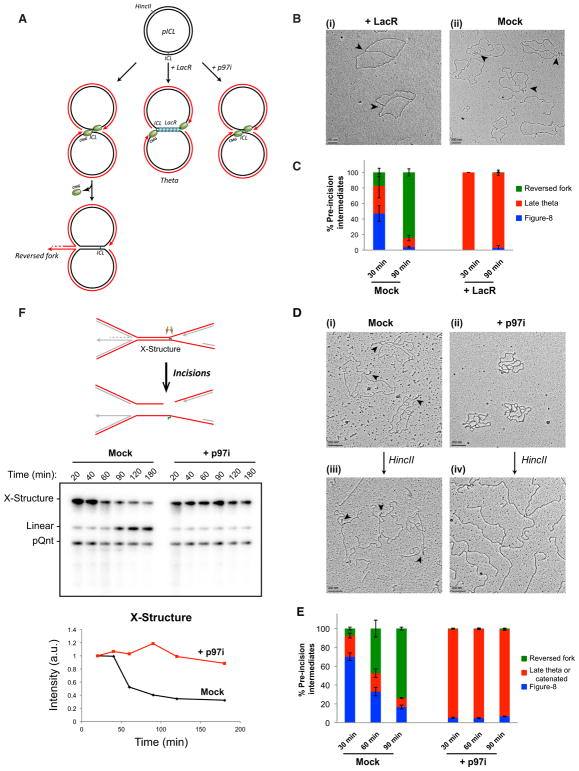
Figure 2. Fork Convergence and CMG Unloading Are Required for Fork Reversal during ICL Repair.
(A) Model depicting replication fork convergence, CMG unloading, and fork reversal in an unperturbed reaction (left), in the presence of lacR (middle), and in the presence of NMS 873 (“p97i”; right).
(B) EM image of late theta structures (black arrows) in a LacR-treated reaction (i) or in a mock (buffer)-treated reaction (ii) at 90 min.
(C) At the indicated times, late theta, figure 8, and reversed-fork structures from the experiment shown in (B) were quantified and graphed. Error bars indicate the range in two independent experiments.
(D) EM images of reversed fork structures or catenated structures in the presence of mock- (i and iii) or p97i-treated (ii and iv) conditions, respectively, before and after HincII digestion. See text for details.
(E) Quantification of late theta, catenated molecules, figure 8s, and reversed forks by EM in a mock (DMSO)-treated or p97i-treated reaction. Error bars indicate the range in two independent experiments.
(F) pICL incision assay in a mock-treated (buffer) or p97i-treated reaction. pICL and an undamaged, internal control plasmid (pQnt) were nick translated with [α-32P] dATP before addition to extracts to induce replication and repair. Repair intermediates were recovered from extract and digested with HincII, separated by denaturing agarose gel electrophoresis, and visualized by autoradiography. Incisions result in loss of the large parental X-shaped structure (red strands in schematic; quantified in graph) and accumulation of a linear species. A similar result was seen in a second, independent experiment.