Abstract
Colon cancer patients receiving chemotherapy continue to be burdened with therapeutic failure and adverse side effects, yielding a need to develop more effective treatments. The present study investigates Cinnamtannin B-1 (CTB-1) as a potential low-toxicity therapeutic alternative for colon cancer. CTB-1-treated DLD-1, COLO 201 and HCT-116 (WT p53 and p53 null) colon cancer cells and CCD 841 CoN normal colon epithelial cells were assessed for changes in survival using MTT assay. The effects of CTB-1 on cell cycle progression and the apoptosis of colon cancer cells were measured using flow cytometry and/or immunofluorescence. The expression profiles of cell survival molecules, particularly apoptotic proteins, in the colon cancer cells were evaluated following CTB-1 treatment via antibody array, then validated by western blot analysis. Additionally, the potential synergy between CTB-1 and 5-fluorouracil (5-FU), a conventional chemotherapeutic agent used in the treatment of colon cancer, against colon cancer cells was assessed using MTT assay and Calcusyn software. The results revealed that CTB-1 signifi-cantly decreased the survival of the DLD-1, COLO 201 and HCT-116 cells in a time and/or dose-dependent manner, with minimal cytotoxicity to normal colon cells. CTB-1 treatment was shown to induce cell cycle arrest and apoptosis of DLD-1 and COLO 201 cells. Of note, CTB-1 modulated the expression of several cell survival molecules, which tend to be deregulated in colon cancer, including p53, a key transcription factor involved in apoptosis. The downstream regulation of Bcl-2 and Bak expression, as well as cytochrome c release into the cytosol, was also observed following CTB-1 treatment. Furthermore, CTB-1 was shown to significantly enhance the potency of 5-FU via a synergistic drug interaction. This study reveals for the first time, to the best of our knowledge, the ability of CTB-1 to decrease the survival of colon cancer cells through pro-apoptotic mechanisms and display synergy with conventional chemotherapy, demonstrating the potential therapeutic benefit of CTB-1 in colon cancer.
Keywords: colon cancer, proanthocyanidins, cinnamtannin B-1, survival, apoptosis, 5-fluorouracil
Introduction
Colon cancer continues to be a major health concern, due to high rates of incidence and mortality (1,2). Current treatments for advanced-stage colon cancer are limited in their capacity to target tumor cells employing adaptive mechanisms necessary for survival, often resulting in the recurrence of disease and a poor prognosis. In addition, the adverse side effects from toxicity associated with conventional treatment approaches continue to hinder the quality of life for vulnerable patients (3–6). Therefore, it is critical to develop novel therapeutic approaches targeting pro-survival mechanisms in colon cancer that are also less toxic.
Conventional chemotherapeutic drugs are commonly used in combination as a neoadjuvant and/or adjuvant treatment for advanced-stage colon cancer, or when surgical resection and radiation are no longer deemed effective treatment options. Several combination chemotherapy regimens may be considered, with each of these regimens commonly consisting of two or three drugs with different mechanisms of action. In the United States, advanced-stage colon cancer patients are generally offered conventional chemotherapies, such as 5-flurouracil (5-FU), oxaliplatin, or a combination of both as a first-line regimen, which has shown success in prolonging patient survival, but still leaves the patient vulnerable to recurrence of the disease (3). In addition, the conventional regimens are also associated with high costs and a multitude of debilitating side effects which can hinder the quality of life of patients (7). Of note, recent studies have demonstrated the use of natural plant products, such as flavonoids, in combination with conventional chemotherapy, as a therapeutic strategy to mitigate toxicity and enhance efficacy (8–11). These findings show the potential for natural plant products to help increase survival and improve the quality of life of colon cancer patients, and provide the basis for further investigation into combinations of other natural agents and conventional chemotherapy in colon cancer.
Proanthocyanidins are naturally occurring polyphenolic bioflavonoids, which are diverse in biological function (12,13). Both in vitro and in vivo experimental data support the anticancer capacity of proanthocyanidins, as they have been shown to reduce survival of tumor cells by inducing cell cycle arrest and apoptosis (14–18). Multiple studies have also revealed the various molecular targets of proanthocyanidins, which could prove to be useful in the prevention or treatment of different cancers (19–22). Although numerous proanthocyanidins have been identified, grape seed proanthocyanidins have been more extensively studied for their anti-cancer effects, as compared with many which have yet to be holistically evaluated in different types of cancer.
Cinnamtannin B-1 (CTB-1) is a naturally occurring trimeric proanthocyanidin, present in a limited number of plants, including Cinnamomum zeylanicum and Laurus nobilis (23,24). CTB-1 has been mostly studied for its ability to inhibit platelet aggregation and potentiate the action of insulin, likely due to its antioxidant properties (25–28). Researchers have also investigated the anti-cancer properties of CTB-1, revealing its cytotoxicity in melanoma cells, and its capacity to induce cell cycle arrest and apoptosis in hepatocellular carcinoma and cervical cancer cells (29,30). Given the observed efficacy of CTB-1 in a select number of cancers, further studies are warranted to determine its efficacy and mechanism of action in other cancers, particularly colon cancer. The current study investigated the anti-survival and pro-apoptotic effects of CTB-1 in colon cancer, while also elucidating cellular and molecular mechanisms underlying CTB-1 function and evaluating the potential for CTB-1 to enhance the potency of conventional chemotherapy. Collectively, these findings, for the first time, at least to the best of our knowledge, implicate CTB-1 as a potential therapeutic alternative to improve colon cancer outcomes.
Materials and methods
Materials
CTB-1, isolated from the L. nobilis plant, was purchased from Enzo Life Sciences (Farmingdale, NY, USA), and was dissolved in DMSO (Corning Life Sciences, Corning, NY, USA). 5-Fluorouracil (5-FU) was purchased from Sigma-Aldrich (St. Louis, MO, USA) and was also dissolved in DMSO. For western blot analysis, p53 rabbit antibody (Ab; cat. no. 2527P), phospho-p53 (Ser6) rabbit Ab (cat. no. 9285P), phospho-p53 (Ser9) rabbit Ab (cat. no. 9288P), Bak (D4E4) rabbit monoclonal antibody (mAb; cat. no. 12105P), cytochrome c rabbit Ab (cat. no. 11940S), GAPDH (D16H11) XP® rabbit mAb (cat. no. 5174S), anti-rabbit IgG HRP-linked Ab (cat. no. 7074P2), and anti-mouse IgG HRP-linked Ab (cat. no. 7076P2) were purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-mouse Bcl-2 mAb (cat. no. 05–826) was purchased from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). Primary antibodies were diluted 1:1,000 and secondary antibodies were diluted 1:2,000 in 5% non-fat milk dissolved in TBS with 0.1% Tween-20. For immunofluorescence, PE-Annexin V Ab (cat. no. 640908) was purchased from Biolegend (San Diego, CA, USA) and used at a concentration of 5 µg/ml. The ProLong™ Gold Antifade Mountant with DAPI for nuclear stain was purchased from Thermo Fisher Scientific, Inc.
Cell lines and cell culture
Normal colon epithelial cells (CCD 841 CoN) and colon cancer cells (DLD-1 from node-positive adenocarcinoma patients; and COLO 201 cells from adenocarcinoma patients with distant metastases) were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). These cell lines were cultured in RPMI-1640 medium (Corning Cellgro, Manassas, VA, USA) supplemented with 10% fetal bovine serum (FBS), 100 µg/ml of streptomycin and 100 U/ml of penicillin (both from HyClone, Pittsburgh, PA, USA). Wild-type p53 and p53-null HCT-116 colorectal carcinoma cells were generously donated by Dr Upender Manne (University of Alabama at Birmingham, Birmingham, AL, USA) and cultured in McCoy's 5A medium (ATCC) supplemented with glutamine, penicillin/streptomycin and 10% FBS. All cells were maintained at 37°C with 5% CO2, and cultured in either RPMI-1640 or McCoy's 5A media with 1% FBS, 24 h prior to each experiment.
Cell viability
The effect of CTB-1, either alone or in combination with 5-FU, on colon cancer cell survival, was assessed by cell viability assay using the tetrazolium dye, MTT. Briefly, normal colon epithelial cells or colon cancer cells (5×104) were cultured in triplicate in a 96-well plate for 24 h at 37°C with 5% CO2. Cells were then treated with increasing concentrations of CTB-1 (10, 20, 50 and 100 µM) or an equivalent dilution of the DMSO vehicle control for 24, 48 and 72 h. For drug combination experiments, cells were treated with increasing concentrations of 5-FU (10, 20 and 40 µM) alone, 5-FU in combination (1:1) with CTB-1 (10, 20 and 40 µM), or an equivalent dilution of the DMSO vehicle control for 72 h. To each well, 20 µl of MTT reagent (0.5 mg/ml in 1X PBS) was added following treatment, followed by incubation for 2 h to allow the formation of insoluble formazan crystals. The media was then discarded, and DMSO was added to dissolve formazan crystals. The optical density was measured at 570 nm using a SpectraMax M5 spectrophotometer (Molecular Devices, San Jose, CA, USA). All concentrations were tested in quadruplicate and experiments were repeated 3 times.
Flow cytometric analysis
To ascertain the effects of CTB-1 on cell cycle progression and apoptosis, DLD-1 and COLO 201 colon cancer cells were treated with CTB-1 (20 or 40 µM) or the vehicle control for 24 h (cell cycle) or 48 h (apoptosis). The range of CTB-1 concentrations was selected based on suboptimal (20 µM) and above-optimal (40 µM) concentrations relative to the half maximal effective concentration (EC50) values determined from the cell viability assay, and the time points based on the inferred sequence and timing of intracellular biological events. For cell cycle analysis, 1×106 cells were harvested and washed 3 times with fluorescence-activated cell-sorting (FACS) buffer (PBS supplemented with 2% FBS). Cells were then stained with propidium iodide containing RNase (Cell Signaling Technology, Danvers, MA, USA) according to the manufacturer's instructions. To assess the populations of apoptotic cells following CTB-1 treatment, cells were harvested and washed in ice-cold PBS. Subsequently, 1×105 cells were stained with PE-Annexin V and 7-aminoactinomycin D (7-AAD) using the PE-Annexin V Apoptosis Detection kit I (BD Biosciences, San Jose, CA, USA), according to the manufacturer's instructions. The fluorescent intensity of the stained cells was acquired (20,000 events/sample) using a Guava flow cytometer (Millipore, Billerica, MA, USA) and analyzed using FlowJo 10.0.06 software (Treestar Inc., Ashland, OR, USA). Apoptotic cells could be grouped into early-stage apoptosis (Annexin V+ and 7-AAD-) and late-stage apoptosis (Annexin V+ and 7-AAD+) shown in the lower right and upper right quadrants of the FACS dot plots, respectively. The experiment was repeated 3 times for both cell cycle and apoptotic analyses.
Immunofluorescence
To determine the effect of CTB-1 on apoptosis, colon cancer cells (DLD-1 and COLO 201) were cultured on Nunc™ Lab-Tek™ II Chamber Slides™ (Thermo Fisher Scientific, Inc.) and then treated with CTB-1 (20 or 40 µM) or the vehicle control for 48 h. Following treatment, cells were washed twice with ice-cold PBS and once with 1X Annexin V Binding Buffer (BD Biosciences). Cells were then incubated with the PE-conjugated Annexin V primary antibody for 20 min. Samples were then mounted on a slide with ProLong™ Gold Antifade Mountant with the nuclear stain DAPI. Images were acquired using Observer.Z1 microscope (Carl Zeiss Microscopy GmbH, Königsallee, Germany).
Antibody array
Antibody microarrays (cat. no. APP069 and PCC076; Full Moon Biosystems, Inc., Sunnyvale, CA, USA) were used to determine the changes in the expression and phosphorylation profile of molecules involved in survival and apoptosis in colon cancer cells (DLD-1 and COLO 201) following CTB-1 treatment. The array layout consisted of highly specific and well characterized antibodies against apoptosis and cell cycle markers, each replicated 6 times, with β-actin and GAPDH as controls. Briefly, colon cancer cells were treated with CTB-1 (40 µM) or DMSO vehicle control for 48 h, and their total protein extracted and biotinylated (75 µg protein/reaction) using an antibody array assay kit (Full Moon Biosystems, Inc.) as per the manufacturer's instructions. Biotinylated samples were coupled with the array slides, which were then labeled with 0.5 mg/ml Cy3-Streptavadin and processed following the manufacturer's instructions. The arrays were scanned and signal quantified by an Axon GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA, USA). For each antibody, the average signal intensity of replicate spots is normalized first to the median signal of the slide, then to the normalized average signal intensity of the GAPDH internal control. Fold changes in protein expression and/or phosphorylation were calculated by dividing normalized average signal intensities for CTB-1 treated samples by untreated controls. Array data was used to generate a heat map using the CIMminer platform (https://discover.nci.nih.gov/cimminer/oneMatrix.do) developed by the Genomics and Bioinformatics Group at the National Cancer Institute.
Isolation of cytoplasmic protein fraction
To determine the effects of CTB-1 treatment on the level of cytosolic cytochrome c, cytoplasmic protein was extracted from DLD-1 and COLO 201 colon cancer cells treated with CTB-1 (20 or 40 µM) or the DMSO vehicle control for 48 h using the NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Briefly, 5×106 cells were harvested, washed and centrifuged at 500 × g for 3 min. Cytoplasmic Extraction Reagent 1 (CER I) was added, followed by vortexing and incubation on ice for 10 min. CER II was then added, followed by vortexing and incubation on ice for 1 min. The cytoplasmic extract was collected by centrifugation at 16,000 × g and stored at -80°C for further analysis.
Western blot analysis
The expression and/or phosphorylation profile of apoptosis related proteins was assessed by western blot analysis. Total protein from colon cancer cells (DLD-1, COLO 201 and HCT-116 WT p53) treated with CTB-1 (20 or 40 µM) or DMSO vehicle control for 48 h were isolated using RIPA lysis buffer containing protease and phosphatase inhibitor cocktail, and quantified by the Pierce bicinconic acid (BCA) method (both from Thermo Fisher Scientific, Inc.). Isolated proteins (30 µg) were resolved on 10% SDS-PAGE and transferred to PVDF membranes using a semidry transfer apparatus (both from Bio-Rad, Hercules, CA, USA). The membranes were incubated in blocking buffer (5% non-fat milk dissolved in Tris-buffered saline with 0.1% Tween-20) for 1 h at room temperature, followed by overnight incubation with primary antibodies at 4°C. Subsequently, the membranes were incubated with secondary antibody for 2 h after washing (3X) with TBST. GAPDH was used as the loading control. Protein bands were developed using Super Signal West Pico Chemiluminescent substrate kit (Thermo Fisher Scientific, Inc.) and images were captured using the ImageQuant LAS 4000 (GE Healthcare Life Sciences, Logan, UT, USA). ImageJ software (http://www.rsbweb.nih.gov/ij) was used to quantify the optical density for treated samples, which were normalized to the GAPDH internal controls.
Evaluating drug interactions
Calcusyn software was used to evaluate drug interactions between CTB-1 and 5-FU, which utilizes the combination index method derived from the median-effect principle established by Chou and Talalay (31–33). The basis of the mathematical model is the median-effect equation fa/fu = [D/Dm]m, where 'fa' is the fraction of cells affected, fu is the fraction unaffected (1-fa), 'D' equals the concentration of the drug, 'Dm' the drug dose required for 50% inhibition, and 'm' is a measurement of the sigmoidicity of the dose-effect curve yielded by Calcusyn. The combination index (CI) is a quantitative representation of the pharmacological interactivity between CTB-1 and 5-FU, which factors in both the potency (Dm) and the shape of the dose-effect curve. CI is derived from the formula CI = (D)1/(Dx)1 + (D)2(Dx)2 + (D)1(D)2/(Dx)1(Dx)2, where (Dx)1 and (Dx)2 are the concentrations for Drug 1 (CTB-1) and Drug 2 (5-FU) separately, resulting in X% inhibition, and (D)1 and (D)2 the concentrations of the respective drugs in combination, resulting in the same percentage inhibition. The CI quantitatively defines synergism as CI<1, additive effect as CI=1 and antagonism as CI>1. The CalcuSyn software generated CI values over a range of Fa levels at different growth inhibition percentages (EC50, EC75 and EC90). The Fa-CI plot, also generated by Calcusyn, is a graphical representation of the pharmacological interaction, which connects the Fa points against the fixed ratio combinations of Drug 1 (CTB-1) and Drug 2 (5-FU) on the X and Y axes. The dose-reduction index (DRI) is a measure of how much the dose of each drug in a synergistic combination may be reduced at a given effect level compared with the doses for each drug alone, and is calculated by the equation (DRI)Drug A = (Dx)Drug A/(D)Drug A.
Statistical analysis
Statistically significant differences between control and treatment groups were determined by one-way ANOVA followed by Dunnett's multiple comparisons test using GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA). A two-way ANOVA followed by Tukey's multiple comparisons test was done to compare statistically significant differences in cell viability between different time points. An unpaired t-test with Welch's correction was used to compare CTB-1 EC50 values in HCT-116 WT p53 and HCT-116 p53 null colon cancer cells. Statistical significance was established at p<0.05 and results are shown as mean ± SEM of 3 independent experiments.
Results
CTB-1 decreases the survival of colon cancer cells
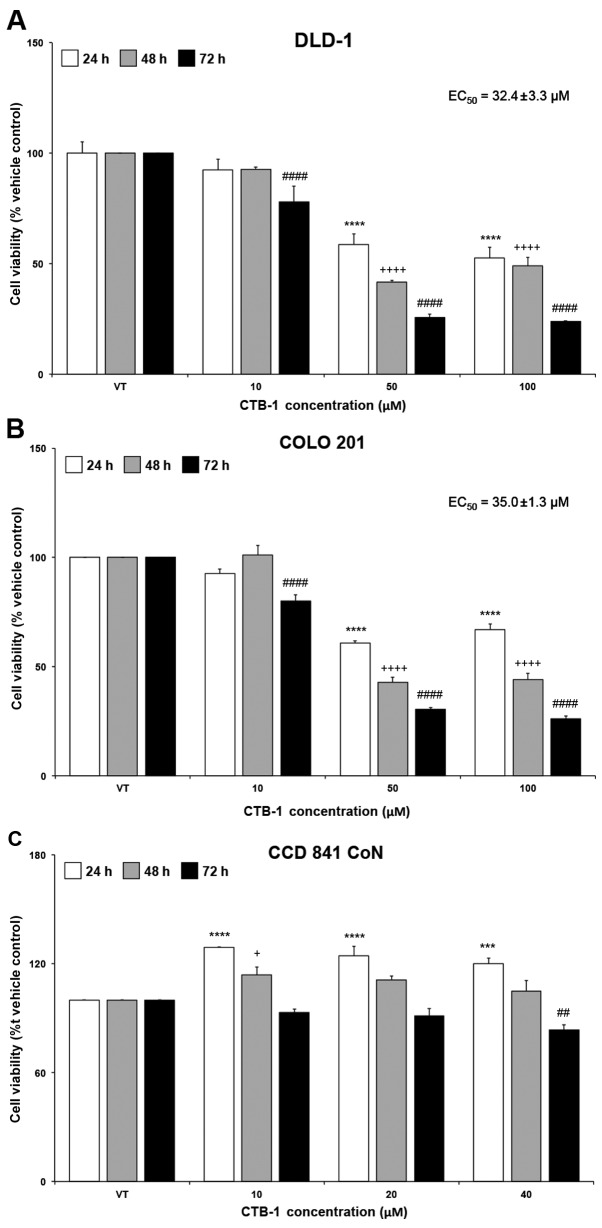
The effect of CTB-1 on the survival of colon cancer cells was assessed using an MTT assay. A dose- and time-dependent reduction in cell viability was observed following treatment with increasing concentrations of CTB-1 in DLD-1 and COLO 201 cells. Compared with the vehicle-treated controls (presented as VT in the figures), a significant reduction was observed after 72 h (DLD-1 cells, 76%, p<0.0001; COLO 201 cells, 74%, p<0.0001) and to a lesser degree at 24 h (DLD-1 cells, 41%, p<0.0001; COLO 201 cells, 39%, p<0.0001) following treatment with CTB-1 (50 µM). Similar time-dependent trends occurred at all concentrations of CTB-1. The EC50 value of CTB-1 was 32.4±3.3 µM in the DLD-1 cells and 35.0±1.3 µM in the COLO 201 cells, calculated from the equation for the line-of-best-fit for the 72-h treatment (Fig. 1A and B). However, minimal effects on the viability of the normal colon cells (CCD 841 CoN) treated within the effective concentration range of CTB-1 (40 µM) were observed (72 h with 40 µM CTB-1 treatment, 16% reduction, p=0.0037), as compared with the vehicle-treated controls (Fig. 1C). These findings highlight the potential of CTB-1 to selectively target colon cancer cells, while possibly decreasing the propensity for adverse side effects from treatment.
Figure 1.
Cinnamtannin B-1 (CTB-1) decreases the viability of colon cancer cells. (A) DLD-1 and (B) COLO 201 colon cancer cell lines were treated with increasing concentrations of CTB-1 or the DMSO vehicle control for 24, 48 and 72 h, then assessed for cell viability via MTT assay. The half maximal effective concentration (EC50) of CTB-1 against colon cancer cell lines following treatment for 72 h is displayed in the graph, calculated from the equation of the line-of-best-fit. (C) CCD 841 CoN normal colon epithelial cells were treated with CTB-1 within the effective concentration range to assess cytotoxicity via MTT assay. For all graphs, the mean value (% vehicle control-VT) ± SEM from 3 replicates is shown. Significant differences between time-matched CTB-1-treated and vehicle-treated samples are denoted by (*) for 24 h, (+) for 48 h, and (#) for 72 h (+p<0.05, ##p<0.01, ***p<0.001, ****,++++,####p<0.0001, Dunnett's multiple comparisons test).
CTB-1 induces cell cycle arrest in colon cancer cells
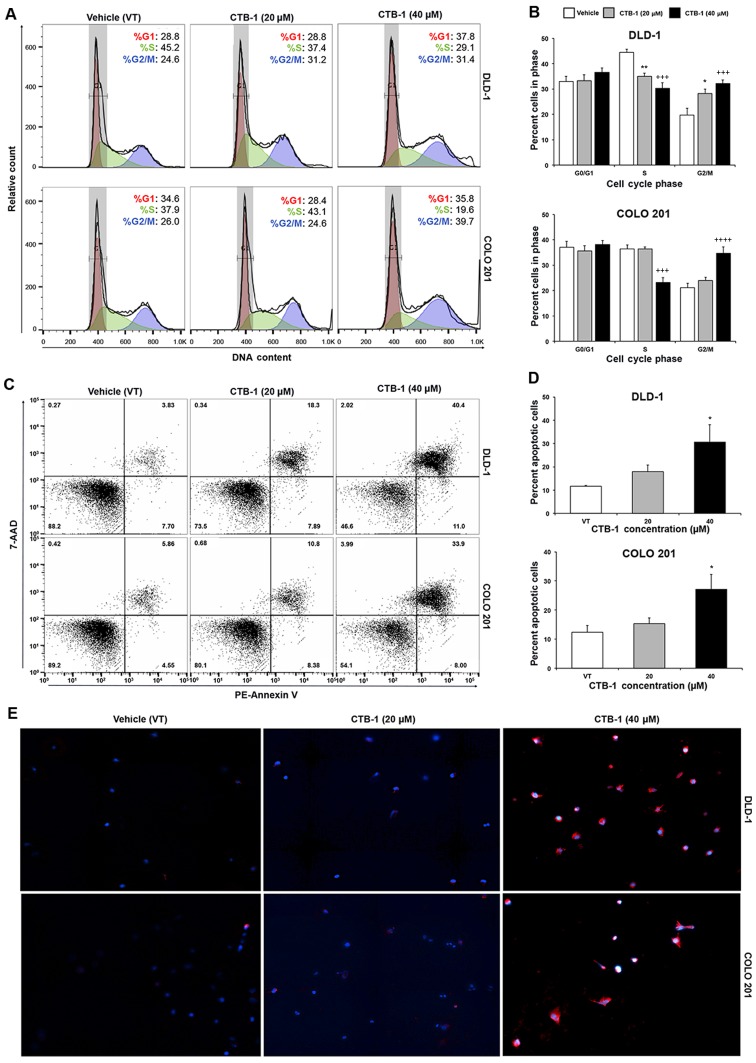
Flow cytometric analysis was performed to ascertain the effects of CTB-1 on the cell cycle phase distribution in colon cancer cells. The DLD-1 (%G2/M: 19.8 to 32.1, p=0.0009) and COLO 201 (%G2/M: 19.8 to 34.7, p=0.0001) colon cancer cells were both significantly arrested in the G2/M phase of the cell cycle following CTB-1 (40 µM) treatment. Our data also revealed that CTB-1 (40 µM) treatment resulted in a significant reduction in the number of DLD-1 (%S: 44.4 to 30.2, p=0.0003) and COLO 201 (%S: 36.4 to 23.2, p=0.0003) cells in the S phase, compared to the vehicle-treated controls (Fig. 2A and B). The ability of CTB-1 to arrest the cell cycle progression of colon cancer cells indeed contributes to the reduction in viable cells observed following treatment, and demonstrates its potential to slow down the growth of tumors.
Figure 2.
Cinnamtannin B-1 (CTB-1) induces cell cycle arrest and apoptosis of colon cancer cells. (A) Representative histograms of flow cytometric analysis of the PI-stained DLD-1 and COLO 201 colon cancer cells. Cells in G0/G1 phase are represented in red; S phase in green; G2/M in blue. (B) Graph representing the percentage of cells in each cell cycle phase following vehicle or CTB-1 treatment. The mean value (% vehicle control-VT) ± SEM from 3 replicates is shown. Significant differences between CTB-1-treated and vehicle-treated samples are denoted by (*) for 20 µM CTB-1 and (+) for 40 µM CTB-1. (*p<0.05, **p<0.01, +++p<0.001, ++++p<0.0001, Dunnett's multiple comparisons test). (C) Representative dot plots of the flow cytometric analysis of the PE-Annexin V-7-AAD double-stained DLD-1 and COLO 201 colon cancer cells. Q1 represents the necrotic cells; Q2 represents the late apoptotic cells; Q3 represents the viable cells; Q4 represents the early apoptotic cells. (D) The percentage of apoptotic colon cancer cells in response to CTB-1. The mean value of total apoptotic cells (% vehicle control-VT) ± SEM from three replicates is shown (*p<0.05, Dunnett's multiple comparisons test). (E) Immunofluorescent PE-Annexin V (red) and DAPI (blue) nuclear staining of colon cancer cells treated with vehicle control or CTB-1.
CTB-1 induces the apoptosis of colon cancer cells
Flow cyto-metric analysis of apoptosis was performed to ascertain whether the reduction in the viability of the colon cancer cells following CTB-1 treatment was due to the induction of apoptosis. Our data revealed an almost 3-fold increase (p<0.05) in the percentage of total apoptotic DLD-1 cells treated with 40 µM CTB-1 (30.6%), as compared with the vehicle-treated controls (11.7%). Similarly, there was >2-fold increase (p<0.05) in the percentage of total apoptotic COLO 201 cells treated with 40 µM CTB-1 (27.1%), as compared with the vehicle-treated controls (12.4%) (Fig. 2C and D). To qualitatively observe the effects of CTB-1 on the apoptosis of colon cancer cells, the DLD-1 and COLO 201 colon cancer cells were treated with CTB-1 (20 or 40 µM) or the vehicle control for 48 h, and then analyzed by immunofluorescence following the PE-Annexin V/DAPI double staining of apoptotic cells. As shown in Fig. 2E, the colon cancer cells treated with CTB-1 exhibited increased PE-Annexin V (red) fluorescent intensity as compared with the vehicle-treated controls. These results indicate that CTB-1 is capable of inducing apoptosis in colon cancer cells.
CTB-1 regulates cell-signaling molecules impacting cell survival and apoptosis
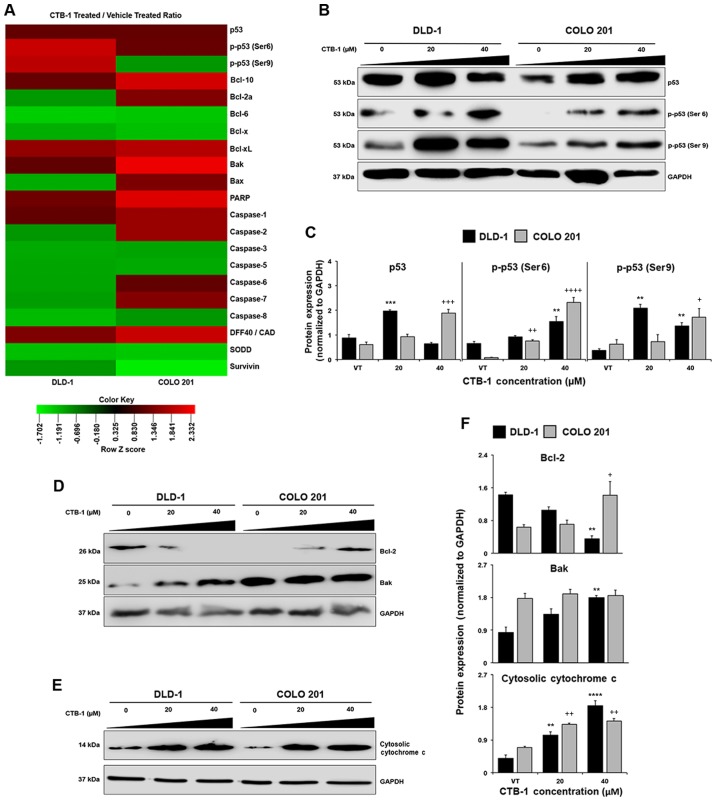
Given the effects of CTB-1 on cell viability and the induction of apoptosis, it was of interest to broadly evaluate proteins that may be regulated and/or activated by CTB-1 using an antibody array. Treatment with CTB-1 (40 µM) was shown to modulate the expression of anti-apoptotic (Bcl-10, Bcl-2a, Bcl-6, Bcl-x and Bcl-xL) and pro-apoptotic (Bak and Bax) Bcl-2 family proteins located on the mitochondrial membrane. Treatment with CTB-1 also modulated the expression of initiator (caspase-2 and -8), effector (caspase-3, -6 and -7) and inflammation-related caspases (caspase-1 and -5), as well as downstream proteins involved in DNA fragmentation (PARP and DFF40/CAD). In addition, CTB-1 modulated the expression of anti-apoptotic proteins involved in the inhibition of extrinsic (SODD) and intrinsic (survivin) apoptotic signaling cascades (Fig. 3A). Most notably, an increase in the phosphorylation of Ser9 (72% increase) and Ser6 (83% increase) p53 residues in the DLD-1 cells was observed following treatment with CTB-1. However, antibody microarray data did not reveal any change in the phosphorylation status of p53 serine residues in CTB-1-treated COLO 201 cells. In addition, there was an increase in the expression of Bak (114% increase) in COLO 201 cells, while no variation was observed in DLD-1 cells. Taken together, these data suggest that CTB-1 may be regulating p53, thereby leading to a modulation of downstream molecules involved in apoptosis.
Figure 3.
Cinnamtannin B-1 (CTB-1) modulates the expression and phosphorylation of apoptotic proteins. DLD-1 and COLO 201 colon cancer cell lines treated with increasing concentrations of CTB-1 or the DMSO vehicle control for 48 h were assessed for fold changes in i) total protein or ii) phosphorylated protein to normal protein. (A) Heatmap representing modulated apoptosis-related proteins included in apoptosis and phospho-specific cell cycle protein antibody arrays. Each row represents one phosphorylation site. Cells shaded red indicate increased expression or phosphorylation and cells shaded green indicate a decrease. (B) Protein expression levels of total p53 and phosphorylation of Ser6 and Ser9 p53 residues were assessed by western blot analysis. (C) Graphs representing the expression of total p53, p53 (Ser6) and p53 (Ser9) based on the optical density (OD) of each protein normalized to the GAPDH control. (D and E) Western blot images of the apoptotic proteins Bcl-2, Bak and cytosolic cytochrome c are shown, as well as (F) graphs representing the expression of each protein based on the optical density of each protein normalized to the GAPDH control. The mean OD value ± SEM from 3 replicates is shown. Significant differences between CTB-1-treated and vehicle-treated samples are denoted by (*) for DLD-1 cells, and (+) for COLO 201 cells (*,+p<0.05, **,++p<0.01, ***,+++p<0.001, ****,++++p<0.0001, Dunnett's multiple comparisons test).
CTB-1 increases the expression and phosphorylation of p53
The results from the antibody arrays led us to validate the expression of p53, as well as the phosphorylation of Ser6 and Ser9 p53 residues by western blot analysis. There was a significant increase (p=0.005) in the expression of total p53 following treatment with 20 µM CTB-1 as compared with the vehicle-treated controls in the DLD-1 cells, yet a decrease was observed at 40 µM CTB-1. There was also a significant increase in the phosphorylation of Ser6 (40 µM CTB-1, p=0.0036) and Ser9 (20 µM CTB-1, p=0.0026; 40 µM CTB-1, p=0.0319) p53 residues following CTB-1 treatment; however, the increase in Ser9 phosphorylation was more pronounced at 20 µM CTB-1 than at 40 µM CTB-1. In the COLO 201 cells, there was a significant dose-dependent increase (40 µM CTB-1, p=0.0002) in total p53, as well as in the phosphorylation of Ser6 (40 µM CTB-1, p=0.0001) and Ser9 (40 µM CTB-1, p=0.02) p53 residues, contrasting from the array data (Fig. 3B and C). The discrepancy in the results of the arrays and western blot analysis were likely due to the maintenance of the tertiary structure for the protein targets in our sample for the array, limiting the number of exposed epitopes to bind to antibody. Our data further revealed that CTB-1 might contribute to the stabilization of p53, and the phosphorylation of activating residues.
CTB-1 regulates the expression of Bcl-2, Bak and the release of cytochrome c into the cytosol
To better understand the downstream molecular consequences following p53 regulation by CTB-1, the expression of key proteins related to apoptosis, namely Bcl-2, Bak and cytochrome c, was evaluated by western blot analysis. In the DLD-1 cells, a significant dose-dependent decrease (40 µM CTB-1, p=0.0049) in the expression of the anti-apoptotic Bcl-2 protein was observed, coupled with a significant increase (p=0.0032) in the expression of the pro-apoptotic Bak protein. Additionally, there was a significant dose-dependent increase (20 µM CTB-1, p=0.0033; 40 µM CTB-1, p=0.0001) in the amount of cytosolic cytochrome c in the CTB-1-treated samples. Of note, in the COLO 201 cells, there was a significant dose-dependent increase (40 µM CTB-1, p=0.02) in the expression of Bcl-2, coupled with no notable change in Bak expression, in contrast to the antibody array results. However, a significant increase (20 µM CTB-1, p=0.0036; 40 µM CTB-1, p=0.0017) in the levels of cytosolic cytochrome c was observed response to CTB-1 (Fig. 3D–F). These findings shed light on the propensity for CTB-1 to induce the initiation of apoptosis by regulating the expression and localization of mitochondrial proteins, possibly through a p53-dependent mechanism.
Loss of p53 mitigates the effect of CTB-1 on colon cancer cells
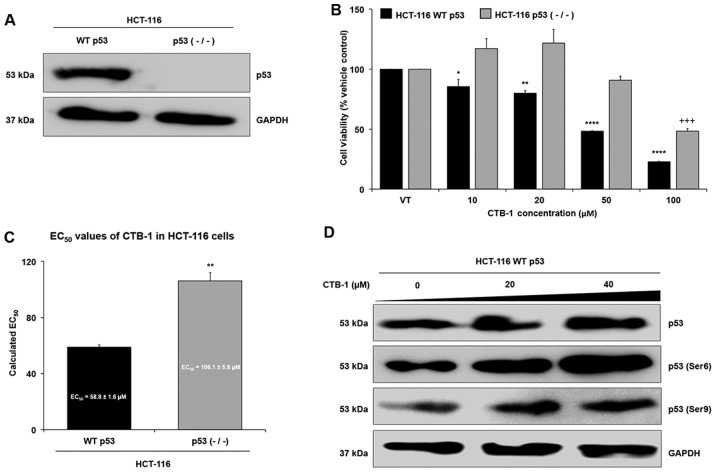
In order to further validate whether CTB-1 exerts its effects on survival via p53, we wished to determine whether CTB-1 similarly decreased the survival and regulated p53 in a colon cancer model with wild-type (WT) p53 rather than the mutated form of p53 in DLD-1 and COLO 201 cells. To do so, HCT-116 colon cancer cells with WT p53, or HCT-116 cells with knockdown of p53 [p53 (−/−)] were compared for the expression of p53 (Fig. 4A) and for the extent to which CTB-1 affected cell viability (Fig. 4B). As shown in Fig. 4C, the EC50 value of CTB-1 (calculated from the equation for the line-of-best-fit for the 72-h treatment) in the HCT-116 WT p53 cells (58.8±1.6 µM) was significantly lower (p=0.003) than the EC50 value of CTB-1 in the HCT-116 p53 null cells (106.1±5.8 µM). In the HCT-116 WT p53 cells, CTB-1 treatment also increased the expression of total p53, and the phosphorylation of Ser6 and Ser9 p53 residues (Fig. 4D), as was observed in the DLD-1 and COLO 201 cells. These data strengthen the notion that CTB-1 may be mediating its effects, in part, via p53.
Figure 4.
Effects of cinnamtannin B-1 (CTB-1) are mitigated by the loss of p53. HCT-116 colon cancer cells with either wild-type (WT) p53 or p53 null (−/−) were (A) validated for p53 status by western blot analysis. (B) WT p53 and p53 null cells were treated with increasing concentrations of CTB-1 or the DMSO vehicle control for 72 h, then assessed for cell viability via MTT assay. Significant differences between CTB-1-treated and vehicle-treated samples are denoted by (*) for WT p53 cells and (+) for p53 null cells. (C) Bar graph representing the half maximal effective concentration (EC50) of CTB-1 against WT p53 and p53 null HCT-116 cells following treatment for 72 h, calculated from the equation of the line-of-best-fit. The mean EC50 value ± SEM from 3 replicates is shown. Significant differences between EC50 values of CTB-1 in p53 null cells versus WT p53 cells are denoted by (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Unpaired t-test with Welch's correction). (D) HCT-116 WT p53 cells were treated with increasing concentrations of CTB-1 or the DMSO vehicle control for 48 h, then protein expression levels of total p53 and phosphorylation at Ser6 and Ser9 p53 residues were assessed by western blot analysis.
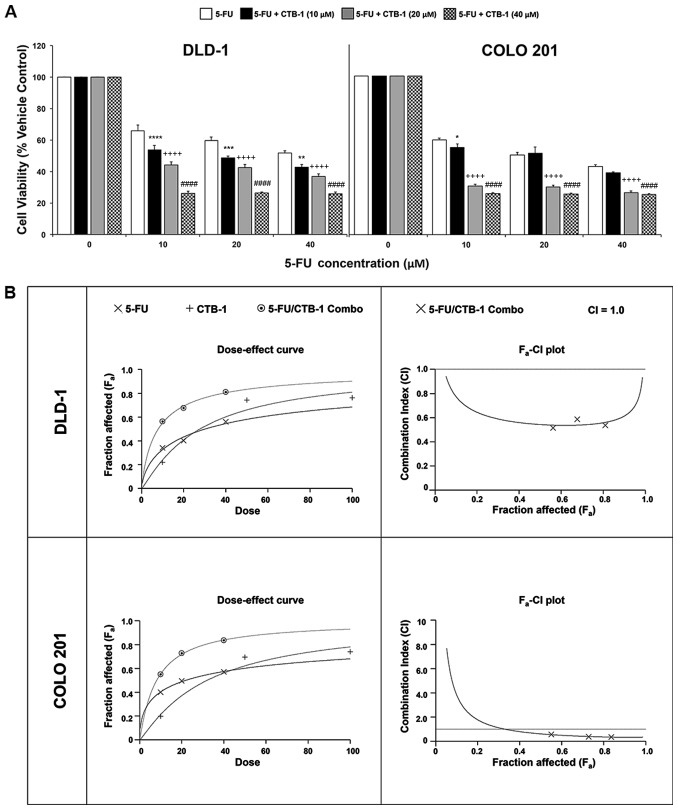
CTB-1 synergizes with 5-FU in exerting its effects on colon cancer cells
To determine whether CTB-1 can function in concert with conventional chemotherapeutic agents, such as 5-FU, to achieve an enhanced potency against colon cancer cells, MTT assay was carried out. Following treatment of the DLD-1 and COLO 201 colon cancer cells with increasing concentrations of 5-FU alone, or in combination with either 10 µM CTB-1, 20 µM CTB-1 or 40 µM CTB-1, viability was significantly lower in cells treated with the combination of 5-FU and CTB-1, as compared with that of cells treated with 5-FU alone (40 µM 5-FU/CTB-1; p=0.0001). In contrast to the DLD-1 cells, in which an enhanced potency was observed with 5-FU treatment in combination with all concentrations of CTB-1, the enhanced potency of the combination treatment in the COLO 201 cells was more prominent in the cells treated with 5-FU and either 20 µM CTB-1 or 40 µM CTB-1 (p=0.0001), as compared with the cells treated with 5-FU and 10 µM CTB-1 (Fig. 5A). To elucidate whether the enhanced effect was due to additive or synergistic drug interactions between 5-FU and CTB-1, Calcusyn software was used to generate dose-effect curves and Fa-CI plots (Fig. 5B). In the dose-effect curve for the DLD-1 and COLO 201 cells, it is shown that the effect of the combination of 5-FU and CTB-1 (1:1 ratio) is greater than the effect of either 5-FU or CTB-1 given alone. The Fa-CI plots for the DLD-1 and COLO 201 cells revealed that the combination index (CI) for 5-FU and CTB-1 (1:1 ratio) at multiple concentrations of both drugs (10, 20 and 40 µM) was below CI=1, indicating a synergistic, rather than additive (CI=1) or antagonistic (CI>1), association between the two drugs. The quantification of the CI at the EC50 (DLD-1, 0.54; COLO 201, 0.62), EC75 (DLD-1, 0.55; COLO 201, 0.39) and EC90 (DLD-1, 0.61; COLO 201, 0.33) of the drug combination is shown in Table I. Additionally in Table I, the Dm, another metric of drug potency, of both 5-FU and CTB-1 is shown to be significantly reduced when given in combination. This increase in drug potency for both 5-FU and CTB-1 was quantified by the drug reduction index (DRI), revealing that in the DLD-1 cells, 5-FU had a DRI=3.85 and CTB-1 had a DRI=3.53 at Fa=0.5. In the COLO 201 cells, 5-FU had a DRI=2.72 and CTB-1 had a DRI=3.99 at Fa=0.5. Overall, these findings indicate there is a true synergistic association between 5-FU and CTB-1 (Table II), indicating CTB-1 may be able to reduce the dosage of 5-FU needed to maintain effectiveness in colon cancer.
Figure 5.
Cinnamtannin B-1 (CTB-1) synergizes with 5-fluorouracil (5-FU) in colon cancer cells. (A) DLD-1 and COLO 201 colon cancer cells were treated with increasing concentrations of 5-FU either alone or in combination with CTB-1 (10, 20 and 40 µM) for 72 h, then assessed for cell viability via MTT assay. The mean value (% vehicle control) ± SEM from 3 replicates is shown. Significant differences between 5-FU/CTB-1-treated samples and samples treated with 5-FU alone are denoted by (*) for 5-FU + CTB-1 (10 µM), (+) for 5-FU + CTB-1 (20 µM), and (#) for 5-FU + CTB-1 (40 µM) (*p<0.05, **p<0.01, ***p<0.001, ****,++++,####p<0.0001, Dunnett's multiple comparisons test). (B) Dose-effect curve and Fa-CI plot generated by Calcusyn software for DLD-1 cells and COLO 201 cells treated with 5-FU alone, CTB-1 alone, or a combination of 5-FU and CTB-1 (1:1 ratio). On the Fa-CI plot, combination index (CI) values below 1 indicate synergism between two drugs.
Table I.
Combination index values for colon cancer cells treated with combination of 5-fluorouracil and cinnamtannin B-1.
| Cell line | Drug | Combination index (CI) values at
|
Dm | DRI (Fa=0.5) | ||
|---|---|---|---|---|---|---|
| ED50 | ED75 | ED90 | ||||
| DLD-1 | 5-FU | – | – | – | 30.20 µM | 3.85 |
| CTB-1 | – | – | – | 27.68 µM | 3.53 | |
| 5-FU + CTB-1 (1:1) | 0.54 | 0.55 | 0.61 | 7.85 µM | – | |
| COLO 201 | 5-FU | – | – | – | 21.80 µM | 2.72 |
| CTB-1 | – | – | – | 31.99 µM | 3.99 | |
| 5-FU + CTB-1 (1:1) | 0.62 | 0.39 | 0.33 | 8.03 µM | – | |
Table II.
Interpretation of combination index values as it relates to additive, synergistic, or antagonistic association between two or more drugs [as described by Chou (33) and analyzed using Calcusyn software].
| Range of CI | Symbol | Description |
|---|---|---|
| <0.1 | +++++ | Very strong synergism |
| 0.1–0.3 | ++++ | Strong synergism |
| 0.3–0.7 | +++ | Synergism |
| 0.7–0.85 | ++ | Moderate synergism |
| 0.85–0.90 | + | Slight synergism |
| 0.90–1.10 | ± | Nearly additive |
| 1.10–1.20 | - | Slight antagonism |
| 1.20–1.45 | -- | Moderate antagonism |
| 1.45–3.3 | --- | Antagonism |
| 3.3–10 | ---- | Strong antagonism |
| >10 | ----- | Very strong antagonism |
Discussion
The majority of colon cancer patients are diagnosed at an advanced stage, at which point chemotherapy often becomes the best, if not only, therapeutic option (34–36). While there have been effective advancements in treatment approaches, from combination therapy to targeted therapy, the fact remains that the 5-year survival rate for advanced stage (stage IIIB-stage IV) patients ranges from 30 to 80%, lower than that of early-stage patients (1). Additionally, the adverse side effects associated with chemotherapy prove to be burdensome. These realities demonstrate room for improvement in therapy, and warrant investigation into alternative treatment approaches, which may simultaneously address issues of efficacy and toxicity.
In the initiation and progression of colon cancer, pro-survival cellular and molecular mechanisms are elevated, allowing cells to divide uncontrollably and avoid apoptosis, normally designed to eliminate abnormal cells (37). As the evasion of apoptosis in advanced-stage colon cancer has been shown to be an important factor in poor response to chemotherapy and radiation, therapies designed to induce apoptosis in target cells would play a critical role in controlling tumor growth (38).
A key cell cycle regulator and apoptosis-related transcription factor is p53, which functions in response to stress and DNA damage in cells. Regarded as 'the guardian of the genome', active p53 causes growth arrest, which provides a window for DNA repair or elimination of cells with severely damaged DNA (39–41). The phosphorylation of serine residues, including Ser6 and Ser9, are essential for the stabilization and activation of p53 (42,43). Once activated, p53 can regulate anti-apoptotic and pro-apoptotic mitochondrial membrane proteins such as Bcl-2 and Bak, respectively, which play a role in the permeabilization of the mitochondrial membrane by opening of voltage-gated anion channels. This consequently leads to the release of cytochrome c from the mitochondria into the cytosol coinciding with the initiation of apoptosis (44). Inactivating mutations in p53 are found in approximately 50% of human cancers, inhibiting its function of transcriptional transactivation of downstream target genes that regulate apoptosis (45).
Our study demonstrates CTB-1, a proanthocyanidin with multiple observed biological functions, can selectively reduce the survival of colon cancer cells (both representative of advanced-stage colon cancer) by approximately 75%, due, to a certain extent, to the induction of apoptosis resulting from regulation of pro- and anti-apoptotic proteins. Of note, the CTB-1-induced increase in the apoptosis of colon cancer cells did not fully correlate with the observed reduction in cell viability, indicating that CTB-1 may be influencing the survival of DLD-1 and COLO 201 cells by alternate cell death mechanisms, such as autophagy. Given the effect of CTB-1 on cell cycle progression, the proliferative capacity of the colon cancer cells may also be reduced in response to treatment, causing the cells to enter cellular senescence. In both p53-mutated DLD-1 and COLO 201 colon cancer cells, CTB-1 treatment resulted in an increased expression of total p53, suggesting that CTB-1 may contribute to the stabilization of p53 within the cells. Furthermore, in response to CTB-1, both cell lines exhibited an increase in the phosphorylation of Ser6 and Ser9, demonstrating that CTB-1 may selectively cause stress in colon cancer cells, and can subsequently restore the activation and function of p53. Although it is possible that the p53 mutations in DLD-1 and COLO 201 cells may have a dominant-negative effect, the fact that CTB-1 treatment demonstrated higher potency against HCT-116 WT p53-expressing cells than HCT-116 p53 null cells, while also increasing the expression of p53 and the phosphorylation of Ser6 and Ser9 residues in HCT-116 WT p53-expressing cells, further demonstrates that CTB-1 may exert its effects, at least in part, through p53-dependent mechanisms.
In this study, while CTB-1 appeared to induce apoptosis by regulating p53 and inducing the release of cytochrome c into the cytosol in both DLD-1 and COLO 201 cells, there were differences observed in the downstream regulation of the mitochondrial proteins, Bcl-2 and Bak, suggesting that p53 may be uniquely interacting with Bcl-2 family proteins in either cell line. One possible reason for the contrasting mechanisms of p53-induced apoptosis may be related to the p53 mutation status, as DLD-1 cells have a missense mutation in p53 (E7 codon 241, Ser to Phe), while COLO 201 cells have a frameshift mutation in p53 (E4 codon 103, T insertion and 25 bp deletion) (46). Since p53 interacts with many pro-apoptotic transcriptional targets, mutations can alter p53 DNA-binding domains via different mechanisms, depending on the cellular context, and can ultimately hinder the ability of p53 to bind to p53-responsive elements on Bcl-2 and Bak genes, or transactivate the transcription of protein inhibitors to Bcl-2, such as PUMA and NOXA (47). As a result, p53 in COLO 201 cells may not have the same capacity as DLD-1 cells to regulate Bcl-2 or Bak expression, and may instead interact with other Bcl-2 family proteins to induce apoptosis. It is also possible that the 48 h time point we selected to evaluate the expression of apoptotic proteins did not coincide with the timing of molecular changes underlying the apop-tosis of COLO 201 cells as it did in the DLD-1 cells. It is also noteworthy that although CTB-1 caused an increase in cytosolic cytochrome c levels in the DLD-1 and COLO 201 cells, the quantification of the mitochondrial membrane potential (MMP) measured via JC-1 assay (data not shown) revealed that CTB-1 treatment did not significantly alter MMP in the colon cancer cell line. The lack of any change in MMP following treatment does not particularly correspond with the regulation of Bcl-2 and Bak mitochondrial proteins and their role in membrane channel assembly. Overall, our findings warrant additional investigation to substantiate the observed reduction in survival.
The idea of using anti-cancer agents together is hardly novel, as the use of combination chemotherapy has yielded increased survival in multiple cancer types and is often standard of care, particularly for advanced-stage cancer patients (3). However, the prospect of finding alternative drug combinations, including ones employing the use of natural plant products, to enhance the potency of conventional chemotherapy is still of great interest, due to the possibility to reduce the required dosage of chemotherapy and the associated adverse side effects from toxicity. Therefore, the ability of CTB-1 to significantly enhance the potency of 5-FU by almost 3–4-fold through synergistic drug interactions highlights the translational/clinical implications that CTB-1 may have by simultaneously combating the high costs and adverse side effects associated with colon cancer treatment, likely leading to improved patient-centered outcomes.
Although our findings establish the anti-survival, pro-apoptotic and synergistic effects of CTB-1 on colon cancer cells in vitro, preclinical studies using a mouse model are required to further strengthen clinical utility of CTB-1 and synergism between CTB-1 and 5-FU.
The data reported in this study add to a growing body of evidence demonstrating the ability of polyphenols, specifically proanthocyanidins, to selectively target cancer cells by inducing apoptosis (14,16–18,20) and to be used in combination with conventional chemotherapy to enhance potency (8–11). Grape seed proanthocyanidins and oligomeric proanthocyanidins have been more extensively studied for their anti-survival properties in multiple cancer types, including colon cancer, and have also been shown to mediate their pro-apoptotic effects via p53 activation (18,21,48,49). By contrast, CTB-1 has been shown to be cytotoxic to melanoma cells (30), and to induce cell cycle arrest and the apoptosis of hepatocellular carcinoma and cervical cancer cells (29); however, it has yet to be holistically evaluated for its anti-cancer properties in colon cancer. The present study, for the first time, at least to the best of our knowledge, revealed CTB-1 increased the expression and phosphorylation of activating p53 residues in both a p53-mutated and WT p53 colon cancer model, which likely precedes the induction of apoptosis and significant reduction in colon cancer cell survival. Given the high frequency of p53 mutations in colon cancer, it is markedly consequential that CTB-1 may restore the p53 function in p53-mutated colon cancer cell lines (50).
Reactive oxygen species (ROS) formation and oxidative stress have been shown to induce neoplastic transformation, as they are involved in several key events of tumorigenesis, including self-sufficiency in growth signals (51,52), and resistance to apoptosis (53,54). The anti-cancer effects of polyphenolic compounds, such as CTB-1, are often attributed to their capacity to scavenge ROS as potent antioxidant molecules. This theory also underlies the ability of polyphenols to target cancer cells over normal cells, as cancer cells may be more sensitive to changes in the redox state, given their increased oxidative stress (55,56). More recently, however, it has been established that polyphenols are also able to induce ROS (mainly superoxide anions or hydrogen peroxide) formation in cancer cells, activating the DNA-damage response pathway (57–61). Since there is evidence to indicate that ROS act as both an upstream signal that triggers p53 activation and as a downstream factor that mediates apoptosis (62–64), it is possible that CTB-1 may influence p53 stabilization, activation and function by affecting ROS levels in colon cancer cells. However, further investigations are warranted to conclude whether this is true, and if so, to determine whether CTB-1 functions as a scavenger or inducer of ROS in colon cancer cells.
In conclusion, in this study, CTB-1 exhibited the capacity to significantly reduce colon cancer survival and induce apoptosis of colon cancer cells, seemingly by modulating the expression of survival-related proteins, specifically those with pro-apoptotic and anti-apoptotic functions, such as p53 and downstream molecular targets. In addition to its anti-cancer efficacy, CTB-1 also exhibited minimal cytotoxicity in normal colon epithelial cells, suggesting a decreased likelihood for adverse side effects. Lastly, CTB-1 displayed a synergistic association with 5-FU, strengthening its clinical implications. Collectively, the data presented in this study taps into the potential of CTB-1 as a novel low-toxicity therapeutic agent for advanced-stage colon cancer.
Acknowledgments
The content of this manuscript benefited from many fruitful conversations with members of the Morehouse School of Medicine, Atlanta, GA, USA. We are also grateful to Dr. Upender Manne from the University of Alabama at Birmingham for generously donating the HCT-116 WT p53 and HCT-116 p53 null colon cancer cell lines for use in our experiments.
Abbreviations
- CTB-1
cinnamtannin B-1
- 5-FU
5-fluorouracil
- VT
vehicle-treated
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphe-nyltetrazolium bromide
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- PE
phycoerythrin
- 7-AAD
7-aminoactinomycin D
- DAPI
4′,6-diamidino-2-phenylindole
- ROS
reactive oxygen species
- RPMI
Roswell Park Memorial Institute
- Ab
antibody
- mAB
monoclonal antibody
- HRP
horseradish peroxidase
Funding
This study was supported in part by the funds (CA169716, CA180212, CA118638 and CA179701) from NCI and Morehouse School of Medicine flow cytometry core. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
PPC developed experimental design, conducted experiments, analyzed data and drafted the manuscript. HM assisted in flow cytometric analysis and manuscript preparation. NK assisted in microscope imaging and manuscript preparation. ABW assisted in western blot analysis and manuscript preparation. SS conceptualized the study, developed study design, supervised the authors throughout the study, and provided his expertise in manuscript preparation. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, Miller D, Bishop K, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, et al. SEER Cancer Statistics Review, 1975–2013. National Cancer Institute; Bethesda, MD: 2016. [Google Scholar]
- 3.Braun MS, Seymour MT. Balancing the efficacy and toxicity of chemotherapy in colorectal cancer. Ther Adv Med Oncol. 2011;3:43–52. doi: 10.1177/1758834010388342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu CY, Chan W, Delclos GP, Du XL. Adjuvant chemotherapy and risk of gastrointestinal, hematologic, and cardiac toxicities in elderly patients with stage III colon cancer. Am J Clin Oncol. 2012;35:228–236. doi: 10.1097/COC.0b013e318210f812. [DOI] [PubMed] [Google Scholar]
- 5.Nadeem H, Jayakrishnan TT, Gamblin TC, Turaga K. Cost differential among systemic therapies for colon cancer. Ann Surg Oncol. 2014;21:S83. [Google Scholar]
- 6.Liu CJ, Lin JK, Chen W-S, Lin TC, Yang SH, Jiang JK, Chang SC, Lan YT, Yen CC, Tzeng CH, et al. The efficacy of chemotherapy in patients with high-grade metastatic colon cancer. Hepatogastroenterology. 2011;58:1495–1501. doi: 10.5754/hge11082. [DOI] [PubMed] [Google Scholar]
- 7.Engstrom PF, Arnoletti JP, Benson AB, III, Chen YJ, Choti MA, Cooper HS, Covey A, Dilawari RA, Early DS, Enzinger PC, et al. National Comprehensive Cancer Network NCCN Clinical Practice Guidelines in Oncology: Colon cancer. J Natl Compr Canc Netw. 2009;7:778–831. doi: 10.6004/jnccn.2009.0056. [DOI] [PubMed] [Google Scholar]
- 8.Patel BB, Sengupta R, Qazi S, Vachhani H, Yu Y, Rishi AK, Majumdar AP. Curcumin enhances the effects of 5-fluorouracil and oxaliplatin in mediating growth inhibition of colon cancer cells by modulating EGFR and IGF-1R. Int J Cancer. 2008;122:267–273. doi: 10.1002/ijc.23097. [DOI] [PubMed] [Google Scholar]
- 9.Patel BB, Gupta D, Elliott AA, Sengupta V, Yu Y, Majumdar AP. Curcumin targets FOLFOX-surviving colon cancer cells via inhibition of EGFRs and IGF-1R. Anticancer Res. 2010;30:319–325. [PMC free article] [PubMed] [Google Scholar]
- 10.Chen WT-L, Yang T-S, Chen H-C, Chen HH, Chiang HC, Lin TC, Yeh CH, Ke TW, Chen JS, Hsiao KH, et al. Effectiveness of a novel herbal agent MB-6 as a potential adjunct to 5-fluoracil-based chemotherapy in colorectal cancer. Nutr Res. 2014;34:585–594. doi: 10.1016/j.nutres.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Amin ARMR, Kucuk O, Khuri FR, Shin DM. Perspectives for cancer prevention with natural compounds. J Clin Oncol. 2009;27:2712–2725. doi: 10.1200/JCO.2008.20.6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plumb GW, De Pascual-Teresa S, Santos-Buelga C, Cheynier V, Williamson G. Antioxidant properties of catechins and proanthocyanidins: Effect of polymerisation, galloylation and glycosylation. Free Radic Res. 1998;29:351–358. doi: 10.1080/10715769800300391. [DOI] [PubMed] [Google Scholar]
- 13.Santos-Buelga C, Scalbert A. Proanthocyanidins and tannin-like compounds - nature, occurrence, dietary intake and effects on nutrition and health. J Sci Food Agric. 2000;80:1094–1117. doi: 10.1002/(SICI)1097-0010(20000515)80:7<1094::AID-JSFA569>3.0.CO;2-1. [DOI] [Google Scholar]
- 14.Prasad R, Vaid M, Katiyar SK. Grape proanthocyanidin inhibit pancreatic cancer cell growth in vitro and in vivo through induction of apoptosis and by targeting the PI3K/Akt pathway. PLoS One. 2012;7:e43064. doi: 10.1371/journal.pone.0043064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma SD, Meeran SM, Katiyar SK. Proanthocyanidins inhibit in vitro and in vivo growth of human non-small cell lung cancer cells by inhibiting the prostaglandin E(2) and prostaglandin E(2) receptors. Mol Cancer Ther. 2010;9:569–580. doi: 10.1158/1535-7163.MCT-09-0638. [DOI] [PubMed] [Google Scholar]
- 16.Singh T, Sharma SD, Katiyar SK. Grape proanthocyanidins induce apoptosis by loss of mitochondrial membrane potential of human non-small cell lung cancer cells in vitro and in vivo. PLoS One. 2011;6:e27444. doi: 10.1371/journal.pone.0027444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nomoto H, Iigo M, Hamada H, Kojima S, Tsuda H. Chemoprevention of colorectal cancer by grape seed proanthocyanidin is accompanied by a decrease in proliferation and increase in apoptosis. Nutr Cancer. 2004;49:81–88. doi: 10.1207/s15327914nc4901_11. [DOI] [PubMed] [Google Scholar]
- 18.Chen Q, Liu X-FF, Zheng P-SS. Grape seed proanthocyanidins (GSPs) inhibit the growth of cervical cancer by inducing apoptosis mediated by the mitochondrial pathway. PLoS One. 2014;9:e107045. doi: 10.1371/journal.pone.0107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaid M, Singh T, Katiyar SK. Grape seed proanthocyanidins inhibit melanoma cell invasiveness by reduction of PGE2 synthesis and reversal of epithelial-to-mesenchymal transition. PLoS One. 2011;6:e21539. doi: 10.1371/journal.pone.0021539. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Engelbrecht AM, Mattheyse M, Ellis B, Loos B, Thomas M, Smith R, Peters S, Smith C, Myburgh K. Proanthocyanidin from grape seeds inactivates the PI3-kinase/PKB pathway and induces apoptosis in a colon cancer cell line. Cancer Lett. 2007;258:144–153. doi: 10.1016/j.canlet.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 21.Nandakumar V, Singh T, Katiyar SK. Multi-targeted prevention and therapy of cancer by proanthocyanidins. Cancer Lett. 2008;269:378–387. doi: 10.1016/j.canlet.2008.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ting-Ting L, Tong L, Yu-Cong Z, Ke-Yuan Z. Inhibitive effect of proanthocyanidins on cyclooxygenase-2 expression in A549 cells induced by cytokine interleukin-1 beta. J Shanghai Jiaotong Univ. 2012;17:500–504. doi: 10.1007/s12204-012-1312-y. [DOI] [Google Scholar]
- 23.Jayaprakasha GK, Ohnishi-Kameyama M, Ono H, Yoshida M, Jaganmohan Rao L. Phenolic constituents in the fruits of Cinnamomum zeylanicum and their antioxidant activity. J Agric Food Chem. 2006;54:1672–1679. doi: 10.1021/jf052736r. [DOI] [PubMed] [Google Scholar]
- 24.Dall'Acqua S, Cervellati R, Speroni E, Costa S, Guerra MC, Stella L, Greco E, Innocenti G. Phytochemical composition and antioxidant activity of Laurus nobilis L. leaf infusion J Med Food. 2009;12:869–876. doi: 10.1089/jmf.2008.0119. [DOI] [PubMed] [Google Scholar]
- 25.Taher M. Majid F adibah abdul and Sarmidi MRS: A proanthocyanidin from cinnamomum zeylanicum stimulates phosphorylation of insulin receptor in 3t3-l1 adipocytes. J Teknol. 2006;44:53–68. [Google Scholar]
- 26.Bouaziz A, Salido S, Linares-Palomino PJ, Sanchez A, Altarejos J, Bartegi A, Salido GM, Rosado JA. Cinnamtannin B-1 from bay wood reduces abnormal intracellular Ca2+ homeostasis and platelet hyperaggregability in type 2 diabetes mellitus patients. Arch Biochem Biophys. 2007;457:235–242. doi: 10.1016/j.abb.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 27.Ben Amor N, Bouaziz A, Romera-Castillo C, Salido S, Linares-Palomino PJ, Bartegi A, Salido GM, Rosado JA. Characterization of the intracellular mechanisms involved in the antiaggregant properties of cinnamtannin B-1 from bay wood in human platelets. J Med Chem. 2007;50:3937–3944. doi: 10.1021/jm070508d. [DOI] [PubMed] [Google Scholar]
- 28.Bouaziz A, Romera-Castillo C, Salido S, Linares-Palomino PJ, Altarejos J, Bartegi A, Rosado JA, Salido GM. Cinnamtannin B-1 from bay wood exhibits antiapoptotic effects in human platelets. Apoptosis. 2007;12:489–498. doi: 10.1007/s10495-006-0014-z. [DOI] [PubMed] [Google Scholar]
- 29.Wen L, You L, Yang X, Yang J, Chen F, Jiang Y, Yang B. Identification of phenolics in litchi and evaluation of anticancer cell proliferation activity and intracellular antioxidant activity. Free Radic Biol Med. 2015;84:171–184. doi: 10.1016/j.freeradbiomed.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 30.Kashiwada Y, Nonaka G, Nishioka I, Chang JJ, Lee KH. Antitumor agents, 129. Tannins and related compounds as selective cytotoxic agents. J Nat Prod. 1992;55:1033–1043. doi: 10.1021/np50086a002. [DOI] [PubMed] [Google Scholar]
- 31.Chou TC, Talalay P. Analysis of combined drug effects: A new look at a very old problem. Trends Pharmacol Sci. 1983;4:450–454. doi: 10.1016/0165-6147(83)90490-X. [DOI] [Google Scholar]
- 32.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 33.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 34.Kelly C, Cassidy J. Chemotherapy in metastatic colorectal cancer. Surg Oncol. 2007;16:65–70. doi: 10.1016/j.suronc.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 35.Chibaudel B, Tournigand C, André T, de Gramont A. Therapeutic strategy in unresectable metastatic colorectal cancer. Ther Adv Med Oncol. 2012;4:75–89. doi: 10.1177/1758834011431592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodwin RA, Asmis TR. Overview of systemic therapy for colorectal cancer. Clin Colon Rectal Surg. 2009;22:251–256. doi: 10.1055/s-0029-1242465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 38.Abraha AM, Ketema EB. Apoptotic pathways as a therapeutic target for colorectal cancer treatment. World J Gastrointest Oncol. 2016;8:583–591. doi: 10.4251/wjgo.v8.i8.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fridman JS, Lowe SW. Control of apoptosis by p53. Oncogene. 2003;22:9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- 40.Haupt S, Berger M, Goldberg Z, Haupt Y. Apoptosis - the p53 network. J Cell Sci. 2003;116:4077–4085. doi: 10.1242/jcs.00739. [DOI] [PubMed] [Google Scholar]
- 41.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 42.Higashimoto Y, Saito S, Tong XH, Hong A, Sakaguchi K, Appella E, Anderson CW. Human p53 is phosphorylated on serines 6 and 9 in response to DNA damage-inducing agents. J Biol Chem. 2000;275:23199–23203. doi: 10.1074/jbc.M002674200. [DOI] [PubMed] [Google Scholar]
- 43.Kaeser MD, Pebernard S, Iggo RD. Regulation of p53 stability and function in HCT116 colon cancer cells. J Biol Chem. 2004;279:7598–7605. doi: 10.1074/jbc.M311732200. [DOI] [PubMed] [Google Scholar]
- 44.Schuler M, Bossy-Wetzel E, Goldstein JC, Fitzgerald P, Green DR. p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J Biol Chem. 2000;275:7337–7342. doi: 10.1074/jbc.275.10.7337. [DOI] [PubMed] [Google Scholar]
- 45.Soussi T, Ishioka C, Claustres M, Béroud C. Locus-specific mutation databases: Pitfalls and good practice based on the p53 experience. Nat Rev Cancer. 2006;6:83–90. doi: 10.1038/nrc1783. [DOI] [PubMed] [Google Scholar]
- 46.Siegel R, Ma J, Zou Z, Jemal A. Cancer Stat. 2014;2014:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 47.Hemann MT, Lowe SW. The p53-Bcl-2 connection. Cell Death Differ. 2006;13:1256–1259. doi: 10.1038/sj.cdd.4401962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al-Suhaibani ES. Antiproliferation and antiactivity of proanthocyanidins against colorectal cancer cells (Caco-2) line through mitochondrial pathway. Int J Adv Sci Tech Res. 2015;4:152–161. [Google Scholar]
- 49.Roy AM, Baliga MS, Elmets CA, Katiyar SK. Grape seed proanthocyanidins induce apoptosis through p53, Bax, and caspase 3 pathways. Neoplasia. 2005;7:24–36. doi: 10.1593/neo.04412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Bodmer WF. Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines. Proc Natl Acad Sci USA. 2006;103:976–981. doi: 10.1073/pnas.0510146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoo MH, Xu XM, Carlson BA, Patterson AD, Gladyshev VN, Hatfield DL. Targeting thioredoxin reductase 1 reduction in cancer cells inhibits self-sufficient growth and DNA replication. PLoS One. 2007;2:e1112. doi: 10.1371/journal.pone.0001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun G, Kemble DJ. To C or not to C: Direct and indirect redox regulation of Src protein tyrosine kinase. Cell Cycle. 2009;8:2353–2355. doi: 10.4161/cc.8.15.9225. [DOI] [PubMed] [Google Scholar]
- 53.Vaughn AE, Deshmukh M. Glucose metabolism inhibits apoptosis in neurons and cancer cells by redox inactivation of cytochrom c. Nat Cell Biol. 2008;10:1477–1483. doi: 10.1038/ncb1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salvioli S, Storci G, Pinti M, Quaglino D, Moretti L, Merlo-Pich M, Lenaz G, Filosa S, Fico A, Bonafè M, et al. Apoptosis-resistant phenotype in HL-60-derived cells HCW-2 is related to changes in expression of stress-induced proteins that impact on redox status and mitochondrial metabolism. Cell Death Differ. 2003;10:163–174. doi: 10.1038/sj.cdd.4401124. [DOI] [PubMed] [Google Scholar]
- 55.Mileo AM, Miccadei S. Polyphenols as modulator of oxidative stress in cancer disease: New therapeutic strategies. Oxid Med Cell Longev. 2016;2016:6475624. doi: 10.1155/2016/6475624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cos P, De Bruyne T, Hermans N, Apers S, Berghe DV, Vlietinck AJ. Proanthocyanidins in health care: Current and new trends. Curr Med Chem. 2004;11:1345–1359. doi: 10.2174/0929867043365288. [DOI] [PubMed] [Google Scholar]
- 57.Lee DH, Lee TH, Jung CH, Kim YH. Wogonin induces apoptosis by activating the AMPK and p53 signaling pathways in human glioblastoma cells. Cell Signal. 2012;24:2216–2225. doi: 10.1016/j.cellsig.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 58.Sharif T, Auger C, Alhosin M, Ebel C, Achour M, Etienne-Selloum N, Fuhrmann G, Bronner C, Schini-Kerth VB. Red wine polyphenols cause growth inhibition and apoptosis in acute lymphoblastic leukaemia cells by inducing a redox-sensitive up-regulation of p73 and down-regulation of UHRF1. Eur J Cancer. 2010;46:983–994. doi: 10.1016/j.ejca.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 59.Shankar S, Srivastava RK. Involvement of Bcl-2 family members, phosphatidylinositol 3'-kinase/AKT and mitochondrial p53 in curcumin (diferulolylmethane)-induced apoptosis in prostate cancer. Int J Oncol. 2007;30:905–918. [PubMed] [Google Scholar]
- 60.Ye R, Goodarzi AA, Kurz EU, Saito S, Higashimoto Y, Lavin MF, Appella E, Anderson CW, Lees-Miller SP. The isoflavonoids genistein and quercetin activate different stress signaling pathways as shown by analysis of site-specific phosphorylation of ATM, p53 and histone H2AX. DNA Repair (Amst) 2004;3:235–244. doi: 10.1016/j.dnarep.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 61.Su CC, Lin JG, Li TM, Chung JG, Yang JS, Ip SW, Lin WC, Chen GW. Curcumin-induced apoptosis of human colon cancer colo 205 cells through the production of ROS, Ca2+ and the activation of caspase-3. Anticancer Res. 2006;26:4379–4389. [PubMed] [Google Scholar]
- 62.Méplan C, Richard MJ, Hainaut P. Redox signalling and transition metals in the control of the p53 pathway. Biochem Pharmacol. 2000;59:25–33. doi: 10.1016/S0006-2952(99)00297-X. [DOI] [PubMed] [Google Scholar]
- 63.Maillet A, Pervaiz S. Redox regulation of p53, redox effectors regulated by p53: A subtle balance. Antioxid Redox Signal. 2012;16:1285–1294. doi: 10.1089/ars.2011.4434. [DOI] [PubMed] [Google Scholar]
- 64.Liu B, Chen Y, St Clair DK. ROS and p53: A versatile partnership. Free Radic Biol Med. 2008;44:1529–1535. doi: 10.1016/j.freeradbiomed.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.