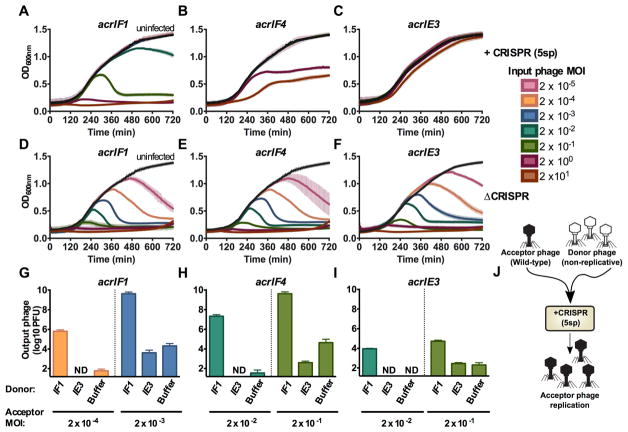
Figure 2. Anti-CRISPR success requires cooperative infections during lytic growth.
(A–F) 12 hour growth curves of P. aeruginosa strain PA14 with 5 targeting spacers (+CRISPR, panels A–C) or no CRISPR-Cas function (ΔCRISPR, D–F) infected with virulent variants of DMS3macrIF1, DMS3macrIF4, or DMS3macrIE3 at multiplicities of infection (MOI) increasing in 10-fold steps from 2×10−5 to 2×101 (rainbow colors) or uninfected (black). Colors correspond to the MOI legend and growth curves. OD600nm is represented as the mean of 3 biological replicates +/− SD (vertical lines). ND, not detectable.
(G–I) Replication of virulent DMS3macr phages (acceptor phage) in the presence of 106 PFU (MOI 0.2) hybrid phage (donor) in PA14 with 5 targeting spacers (5sp) expressing the JBD30 C repressor. Phages were harvested after 24 hours of co-culture and DMS3macr phage PFUs were quantified on PA14 0sp expressing the JBD30 C repressor. Phage output is represented as the mean of 3 biological replicates +/− SD. ND, not detectable.
(J) Schematic of the experimental design in G–I, where a high MOI of non-replicative “donor” phages is used to rescue a low MOI infection of wild-type “acceptor” phages.