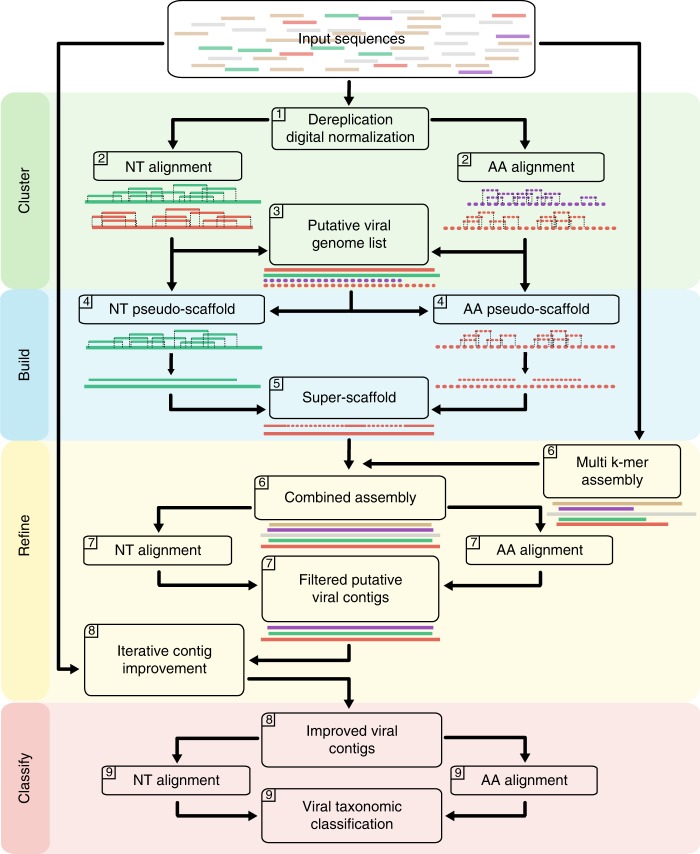
Fig. 1.
A schematic overview of VirMAP. Data processing with VirMAP is achieved through four main stages (shaded colors) divided into nine major steps (top left corner). A putative list of viral genomes and protein pseudo-scaffolds are constructed from clustered nucleotide and translated alignments to the Genbank viral and phage divisions (gbvrl and gbphage). Nucleotide and amino acid pseudo-scaffolds are “built” and merged into a single super-scaffold per genome. A merged de novo assembly is constructed and merged in, resulting in contigs that are then refined using an iterative rebuild process. The improved dual assembly is filtered against a comprehensive Genbank database and are taxonomically classified using a novel per-base contig scoring system