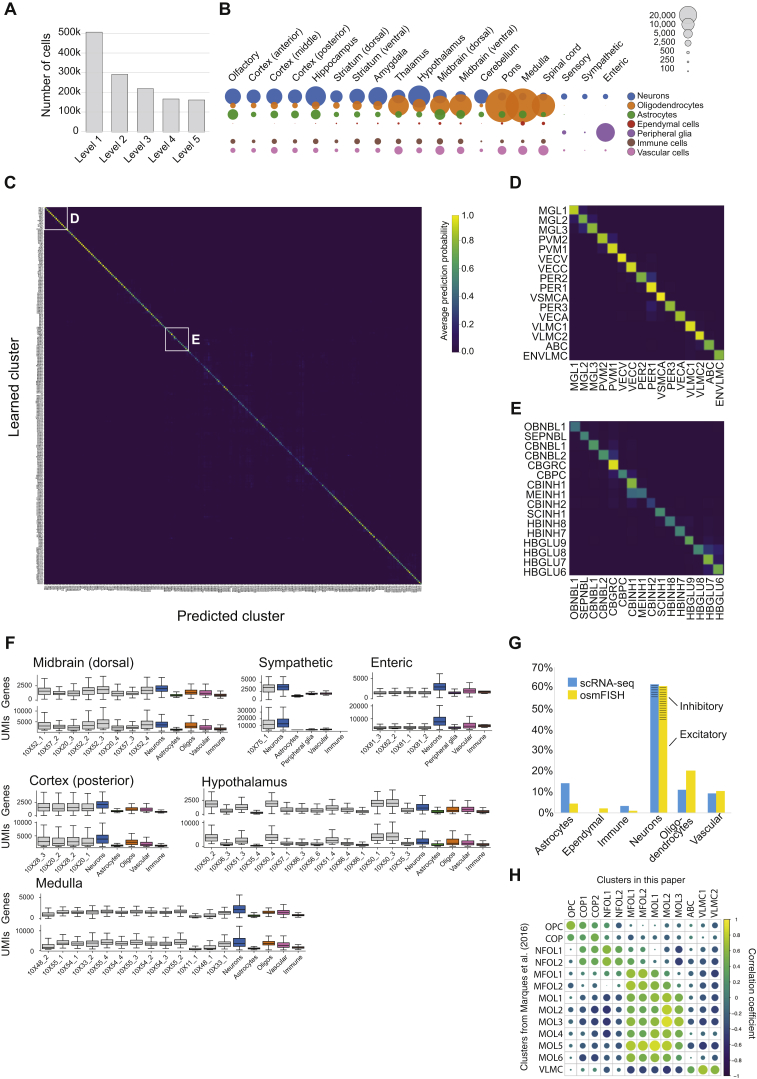
Figure S1.
Data Quality, Related to Figure 1
(A) Number of cells retained in analysis for each level of the pipeline.
(B) Circle plots showing number of cells from each main class and each dissection region.
(C) Cluster robustness and relatedness. The heatmap illustrates the performance of a random forest classifier, showing the average probability assigned to every cell type (rows) for each test cell of given type (columns). When the correct cell type (diagonal) has high probability, almost every test cell will be correctly classified.
(D,E) Magnified view of heatmap as indicated in (C).
(F) Distribution of Gene and UMI counts for individual Chromium samples (gray) and major cell classes (colored), shown for each of a representative selection of tissues.
(G) Comparison of cell type fractions observed by osmFISH (single-molecule fluorescent in situ hybridization) and scRNA-seq.
(H) Comparison of oligodendrocyte lineage clustering in the present paper and those previously published in Marques et al., 2016.