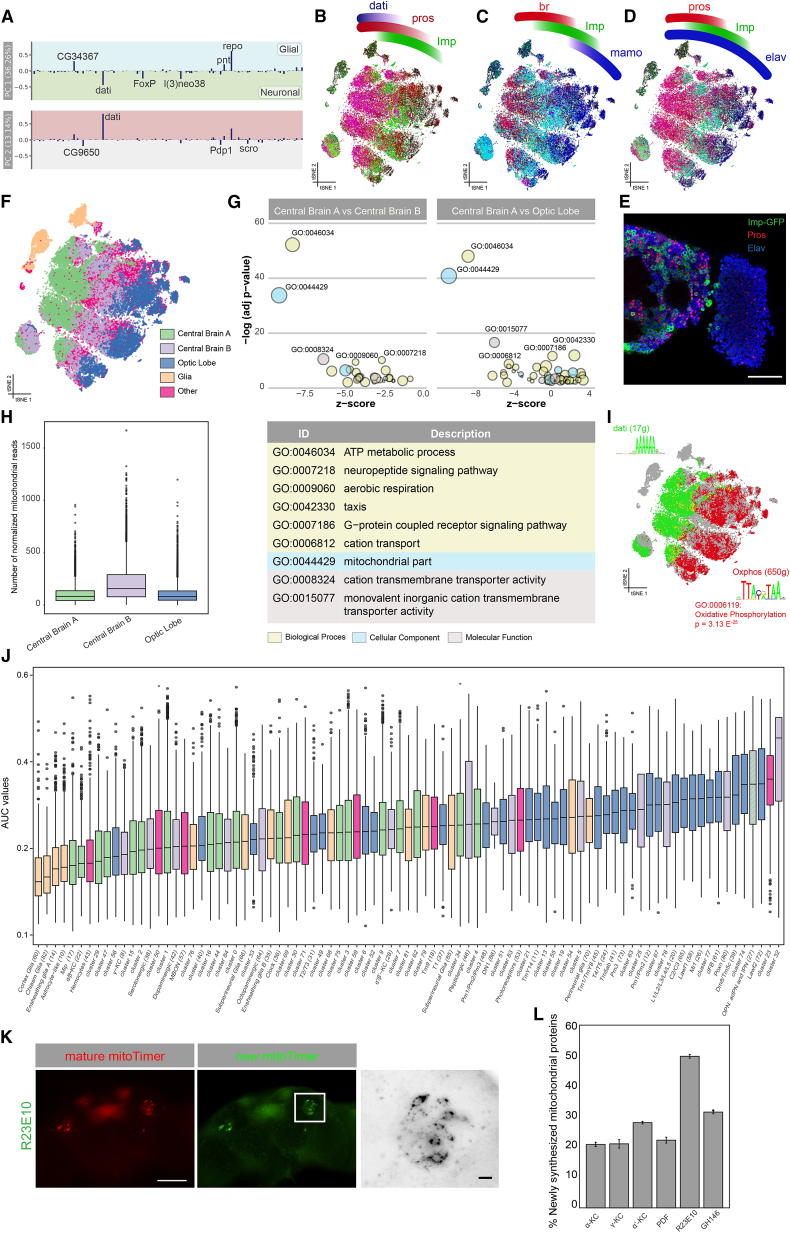
Figure 4.
SCENIC Uncovers a Neuronal Axis Guided by Oxidative Phosphorylation
(A) Loadings for the top two principal components for each regulon.
(B–D) SCENIC t-SNE showing expression of neuroblast “master regulators.”
(E) Immunostaining of Pros and Imp-GFP showing a very strong disjoint expression of both factors, in agreement with the t-SNE.
(F) Division of the t-SNE into three main parts based on expression of dati and Imp: central brain A (dati high), central brain B (dati low, Imp positive), and optic lobe (dati low). Glia were grouped separately, as were neurons that did not meet any of these thresholds.
(G) GO analysis of differential expression between the three main neuronal groups reveals metabolic terms related to oxidative phosphorylation to be downregulated in central brain A.
(H) Number of mitochondrial reads in each neuronal group showing an enrichment in central brain B and optic lobe.
(I) SCENIC t-SNE showing non-overlapping expression of the Dati regulon (green) and a regulon enriched for oxidative phosphorylation (red). Motifs for both regulon are shown, where the OxPhos-enriched motif is the same as previously described in literature.
(J) Boxplot showing the OxPhos expression measured by AUCell for all Seurat clusters. Clusters are colored by which compartment they are part of.
(K) MitoTimer measurements on the R23E10-Gal4 dFBs, showing newly synthesized mitochondrial proteins in green and matured mitochondrial protein in red.
(L) Percentage of newly synthesized mitochondrial proteins for six neuronal populations, colored by central brain compartment. Error bars show 95% confidence interval. Scale bars, 50 μm in half-brain images, 5 μm in zoom-ins.
See also Figure S5.