Figure 3.
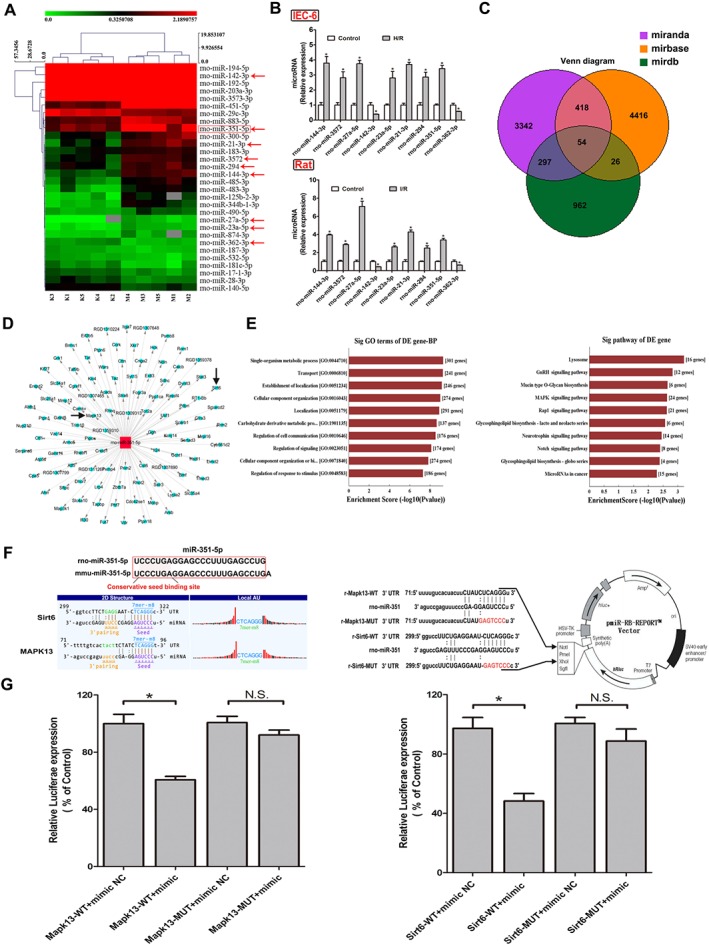
The differentially expressed miRNAs from intestinal tissue of rats after II/R injury based on microarrays assay. (A) Hierarchical clustering analysis of the differentially expressed miRNAs (changes >1.5‐fold and P < 0.05) between II/R and control groups in rats (n = 5). (B) Validation of the nine differentially expressed miRNAs in rats and IEC‐6 cells by real‐time PCR assay (n = 5). (C) Diagram of the search for the target genes of miRNAs in three databases. (D) The target genes of miR‐351‐5p screened from the database. (E) Biological processes and signal pathways of the target genes of miR‐351‐5p. (F) Diagram of miR‐351‐5p conservative seed binding sites on the 3′UTRs of the target genes (Sirt6 and MAPK13) and the construction diagram of double luciferase reporter genes (The mutant sequences used in double luciferase reporter genes are in red). (G) The relative luciferase expression with Sirt6 3′UTR or MAPK13 3′UTR after co‐transfection with miR‐351‐5p mimic or NC in IEC‐6 cells (n = 5). All data are expressed as the mean ± SD. * P < 0.05, significantly different from control group.
