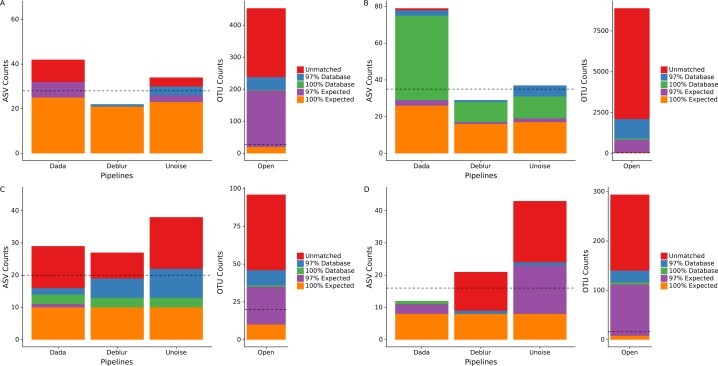
Figure 1. Total number of ASVs/OTUs identified by each sequence processing method for four different mock communities.
Amplicon sequence variants/Operational taxonomic units (ASVs/OTUs) were compared to a database of full-length amplicon sequences for just the microbes supposedly in the community (“Expected”) and against the full SILVA or ITS databases (“Database”) using BLASTN at 97% and 100% identity cut-offs. “Unmatched” sequences did not match an expected sequence or the SILVA/ITS databases at 97% identity or greater. Dotted lines indicate the total number of ASVs/OTUs expected, accounting for 16S copy variation within genomes. Note that the y-axis for open-reference OTU clustering is different than the y-axis on the denoising methods. (A) Human Microbiome Project equal abundance mock community; (B) Extreme dataset; (C) Fungal ITS1 mock community; (D) Zymomock community.