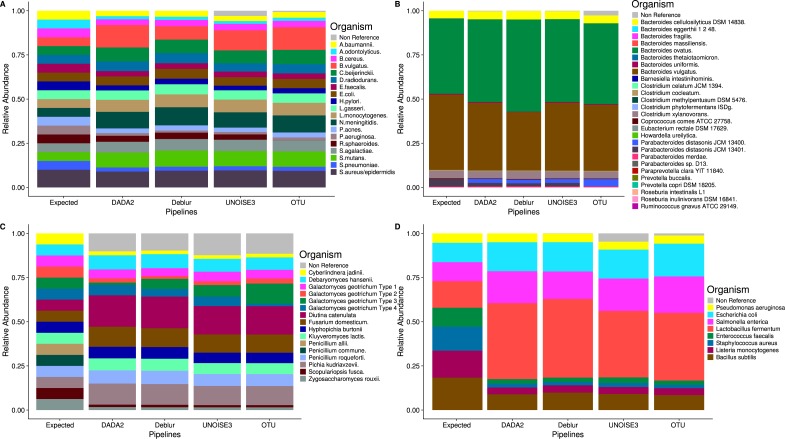
Figure 2. Relative abundances of taxa generated by each sequence processing method for four different mock communities.
All ASVs/OTUs that matched with expected sequences at 97% or greater identity were assigned taxonomy using a BLASTN search against the expected sequences provided for the Extreme, Human Microbiome Project, and Zymomock mock communities. All ASVs/OTUs that matched an expected species with 97% or greater identity to the UNITE database were classified as expected sequences in the fungal mock community. Non-reference refers to the abundance of ASVs/OTUs that did not match expected sequences with 97% or greater identity. (A) Human Microbiome Project equal abundance mock community; (B) Extreme dataset—it is important to note that some organisms are not displayed in this figure due to their very low abundances; (C) Fungal ITS1 mock community; (D) Zymomock community.