Figure 1.
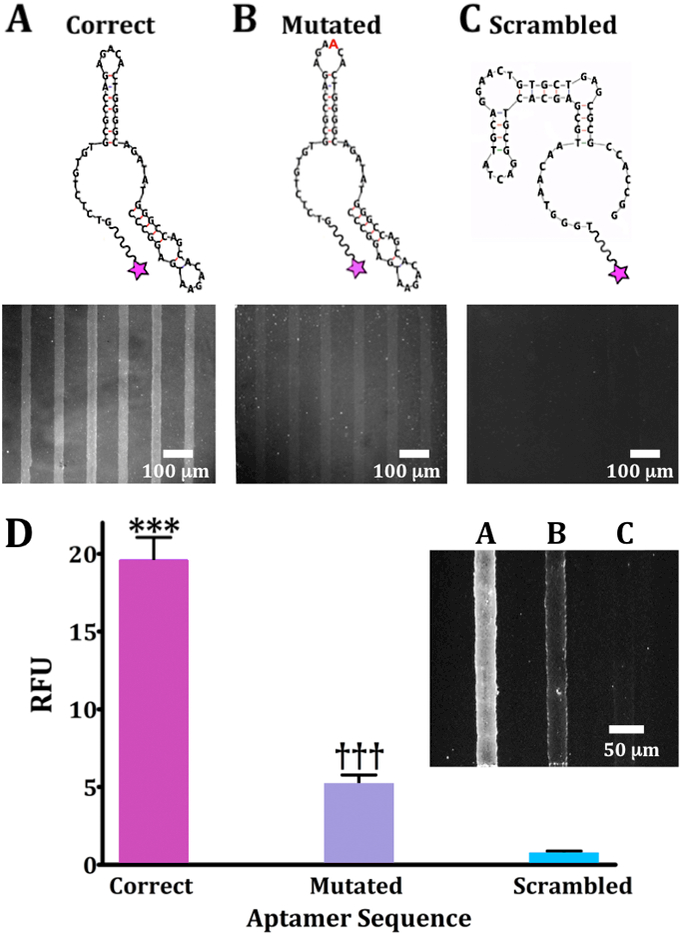
Oligonucleotide binding on dopamine-patterned substrates. Representative fluorescence images are from N=3 substrates per sequence for (A) a 57-base correct dopamine aptamer sequence, (B) a mutated sequence with an extra adenine (red) in a recognition loop at position 21, and (C) a scrambled sequence with the same nucleotides as the correct sequence but a different primary sequence to generate a different secondary structure. Substrates were imaged at an emission wavelength of 605 nm for AlexaFluor® 546 (excitation at 556 nm). (D) Relative fluorescence intensities for patterned dopamine substrates exposed to the three aptamer sequences (20 μM) captured on the same substrate but in different channels (inset; representative image). Error bars are standard errors of the means for N=3 substrates. Group means were significantly different [F(2,15)=719; P<0.001]; ***P<0.001 vs. mutated or scrambled; †††P<0.001 vs. correct or scrambled.
