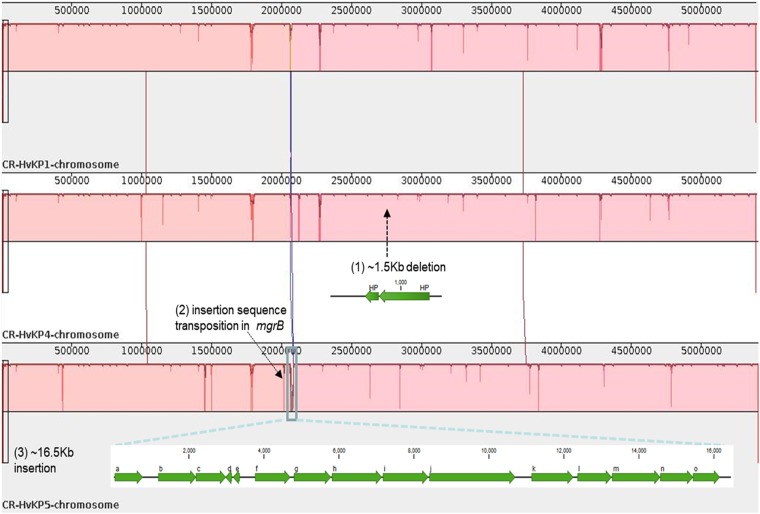
Fig. 1. Comparison of the ST11 CR-hvKP genomes.
Major regions of divergence are labeled, including: (1) An 1.5-Kb deletion in the K. pneumoniae 4 genome with two genes encoding hypothetical proteins (indicated with a black arrow); (2) disruption of the mgrB gene by insertion sequence (ISKpn18) in the K. pneumoniae 5 genome (indicated with a black arrow); (3) A 16.5-Kb region of the K. pneumoniae 5 genome (indicated by a blue rectangle) is absent in the other two ST11 K. pneumoniae genomes. Predicted genes within this region are indicated: (a) peptidase; (b) fatty acid desaturase; (c) phosphate ABC transporter substrate-binding protein; (d) nitrilotriacetate monooxygenase; (e) hypothetical protein; (f) transcriptional regulator TdcA; (g) bifunctional threonine ammonia-lyase/L-serine ammonia-lyase TdcB; (h) threonine/serine transporter TdcC; (i) propionate kinase; (j) formate C-acetyltransferase; (k) branched-chain amino acid ABC transporter substrate-binding protein; (l) branched-chain amino acid ABC transporter permease, LivH; (m) branched-chain amino acid ABC transporter permease, LivM; (n) ABC transporter ATP-binding protein, LivG; (o) ABC transporter ATP-binding protein, LivF