Fig. 4.
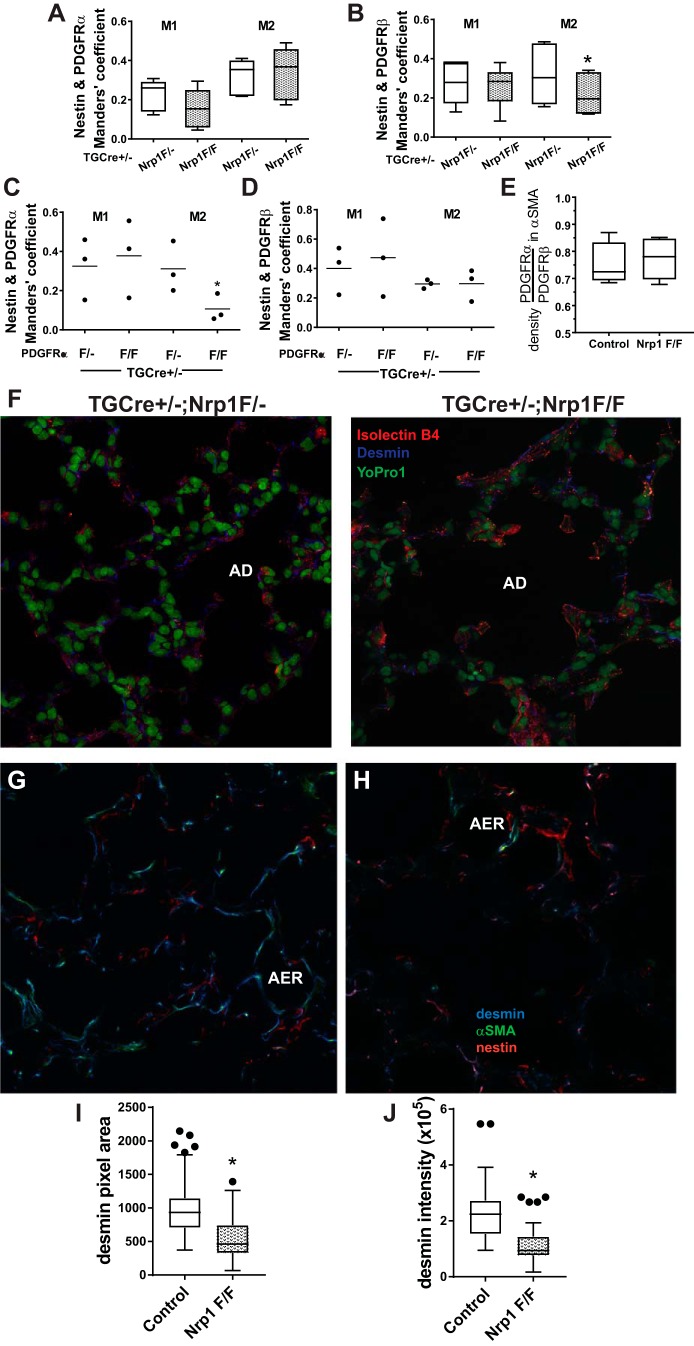
Nestin (NES) and desmin (DES) are differentially distributed in mesenchymal cells after neuropilin-1 (nrp1) gene deletion. A−H: at postnatal day 12 (P12), lungs from TGCre+/−;Nrp1F/F mice and littermate TGCre+/−;Nrp1F/− controls were uniformly inflated and fixed, and 7-μm frozen sections were prepared. Tissues were immunostained for platelet-derived growth factor (PDGF)-α receptor (PDGFRα), PDGFRβ, and NES and imaged using confocal microscopy with uniform parameters within staining and imaging sessions, which always included both paired littermates. Consistent thresholding criteria and the ImageJ2 JACoP plugin were used to calculate Mander’s coefficients for the proportion of pixels occupied by one of the PDGFRs (PDGFRα or PDGFRβ) that overlap pixels occupied by NES (M1) or the proportion of pixels occupied by NES that overlap pixels occupied by one of the PDGFRs (M2). Box-and-whisker (Tukey) plots show data from lungs of 5 separate mice for each genotype; Manders’ coefficients for TGCre+/−;Nrp1F/F mice are compared with those of their respective TGCre+/−;Nrp1F/− littermate mice. *P < 0.01 for comparison of M2 for NES within PDGFRβ (by 2-way ANOVA). C and D: lungs from 3 TGCre+/−;PDGFRαF/F and 3 TGCre+/−;PDGFRαF/− littermate control mice were prepared and analyzed as described in A. Manders’ coefficients for TGCre+/−;PDGFRα1F/F mice were compared with controls. *P < 0.05 for comparison of M2 for NES within PDGFRα. E: sections prepared from the same mice used for experiments described in A and B were stained for α-smooth muscle actin (α-SMA), PDGFRα, and PDGFRβ. Consistent criteria were used to threshold pixels containing α-SMA, and intensities of PDGFRα or PDGFRβ within α-SMA-containing pixels were ascertained. Data are presented as the ratio of PDGFRα intensity to PDGFRβ intensity within α-SMA-containing pixels. Box-and-whisker (Tukey) plots show data from 5 separate mice for each genotype. F: sections of lungs from TGCre+/−;Nrp1F/− and TGCre+/−;Np1F/F mice at P21 were stained for DES, the endothelial marker isolectin B4, and the nuclear stain YO-PRO-1 and imaged using confocal microscopy. Images are representative of those obtained from 4 fields using sections prepared from 3 mice of each genotype. AD, alveolar duct. G−J: images of confocal z stacks from 100-μm sections from 5 TGCre+/−;Nrp1F/F and 5 littermate TGCre+/−;Nrp1F/− control mice immunostained for DES, NES, and α-SMA. For image analysis, uniform thresholding and segmentation criteria were applied to compare nrp1-deleted and control lungs analyzed within the same staining and imaging session. G and H: representative images for lungs from control (G) and nrp1-deleted (H) mice at z levels that contain an alveolar entry ring (AER). Pixel areas and integrated intensities of pixels occupied by DES were ascertained for all 5 littermate pairs, obtained from 4 separate litters. I and J: areas and intensities were pooled for all AERs analyzed for each genotype (n = 73 TGCre+/−;Nrp1F/− and n = 56 TGCre+/−;Nrp1F/F). *P < 0.05 (by Student’s t-test for unpaired variables and Bonferroni’s correction for multiple comparisons). ●, Outliers.