FIGURE 3.
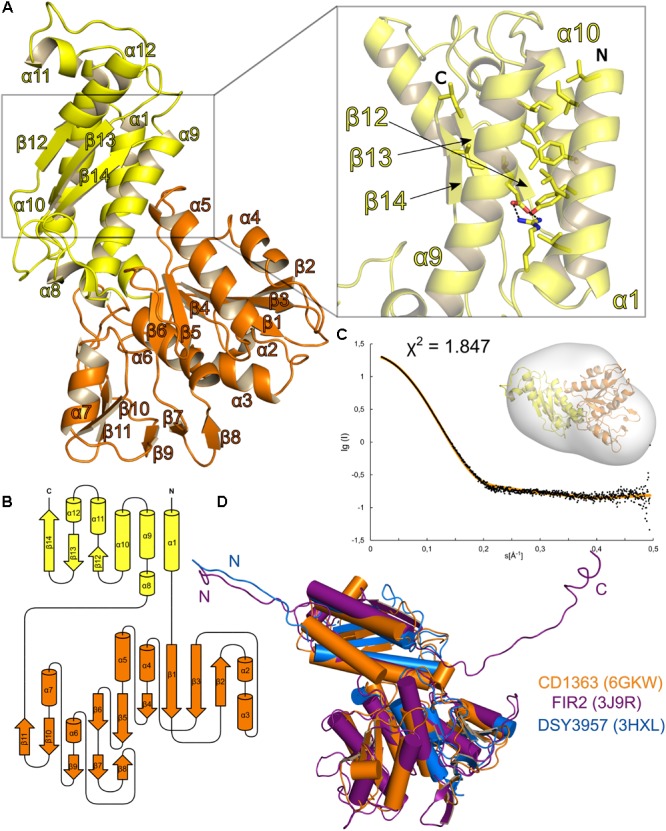
Structural analysis of Clostridium difficile R-type diffocin sheath protein CD1363. The structure of monomeric CD1363 is shown as cartoon representation in (A) with Domain I and II colored in yellow and orange, respectively. Domain I contains a central three-helix bundle that is stabilized by hydrophobic interactions and by a salt bridge (see insert). The C-terminal part of this domain is organized as a three-stranded β-sheet that hydrophobically interacts with the helix bundle. The general domain organization of CD1363 is shown as topology diagram in (B). SAXS analysis of purified CD1363 is presented in (C) with a fit of the experimental SAXS data (black dots) and a theoretical curve calculated from the crystal structure (orange dots) as well as a fit of the CD1364 crystal structure to the SAXS envelope. In (D), monomeric structures of homologous sheath proteins from assembled R-type pyocin (FIIR2; PDB: 3J9R; Ge et al., 2015) and from monomeric DSY3957 from Desulfitobacterium hafniense (PDB: 3HXL; Aksyuk et al., 2011) were superimposed onto CD1363. N- and C-termini are labeled with N and C, respectively.
