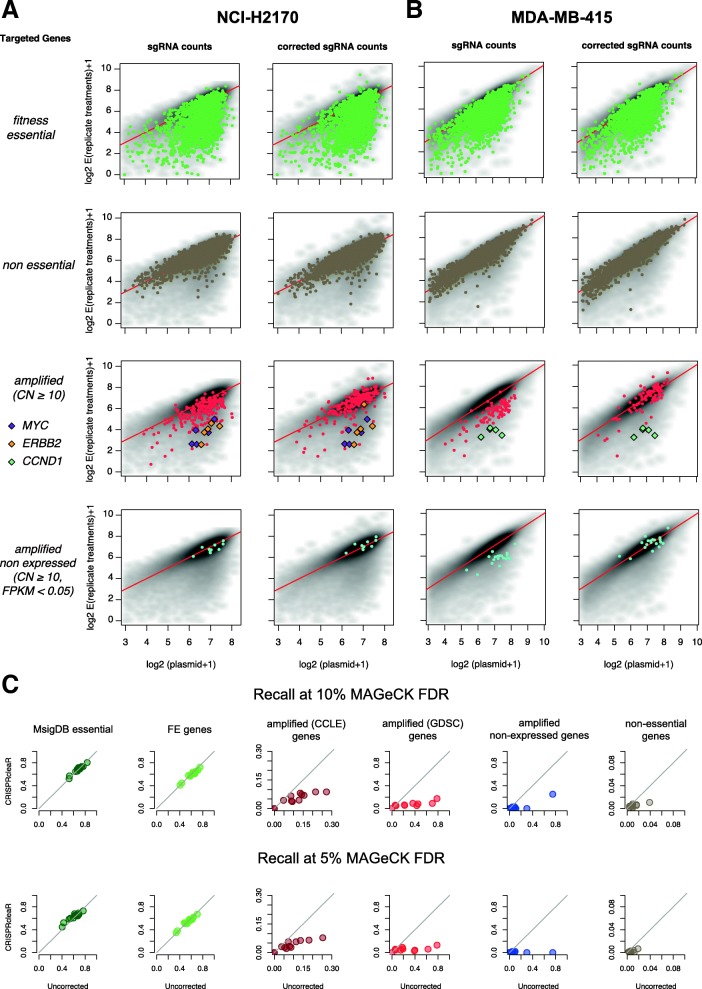
Fig. 5.
CRISPRcleanR corrected sgRNA counts and downstream analysis with MAGeCK. a and b Normalised counts of sgRNAs of the transfected libraries versus the control plasmid for FE and non-essential genes (first two rows of plots), CN amplified genes (third row) and CN non-expressed genes (fourth row), for two example cell lines before (first and third column) and after (second and fourth column) CRISPRcleanR correction. Essentialities for CN-amplified cancer driver genes such as MYC, ERBB2 and CCND1 are retained post correction. For the sake of readability only genes with at least 10 copies have been highlighted. c Comparison of recall using MAGeCK for sgRNAs targeting genes in six predefined gene sets when using as input CRISPRcleanR uncorrected and corrected sgRNAs counts