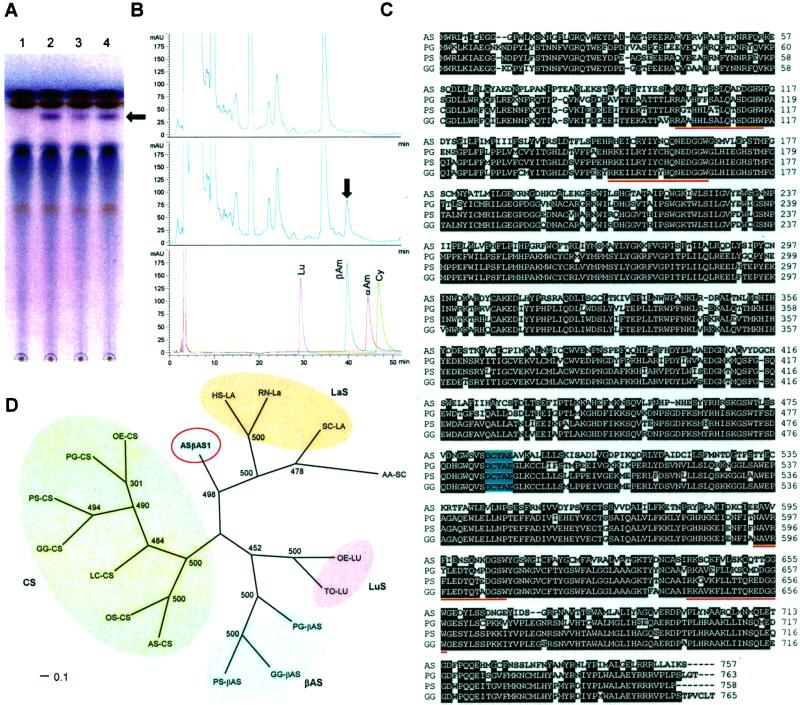
Figure 1.
Characterization of A. strigosa β-amyrin synthase. (A and B) Expression in yeast. (A) TLC analysis of yeast cell extracts. Lane 1, expression vector alone; lanes 2 and 3, expression vector containing full-length A. strigosa β-amyrin synthase cDNA clones (with 59 and 36 bp of 5′ untranslated leader sequence, respectively); and lane 4, P. ginseng β-amyrin synthase cDNA clone pOSCPNY (8). The migration position of a β-amyrin standard is indicated by the arrow. (B) HPLC analysis of yeast cell extracts. (Top) Expression vector alone; (Middle) expression vector containing the A. strigosa β-amyrin synthase cDNA; (Bottom) standards: Lu, lupeol; βAm, β-amyrin; αAm, α-amyrin; Cy, cycloartenol. (C) Alignment of the deduced amino acid sequence of AsbAS1 with those of other β-amyrin synthase genes. PG, P. ginseng (GenBank accession no. AB009030); PS, P. sativum (GenBank accession no. AB034802); GG, Glycyrrhiza glabra (GenBank accession no. AB037203). Conserved amino acid residues are boxed in black, the QW motifs are underlined in red, and the DCTAE substrate-binding site is indicated in blue. (D) Amino acid sequence relatedness of AsbAS1 and other members of the oxidosqualene cyclase superfamily. ASβAS, A. strigosa AsbAS1 (GenBank accession no. AJ311789); LaS, lanosterol synthases: HS-LA, Homo sapiens (GenBank accession no. U22526); RN-LA, Rattus norvegicus (GenBank accession no. U31352); SC-LA, Saccharomyces cerevisiae (GenBank accession no. U04841); AA-SC, Alicyclobacillus acidocaldarius squalene-hopene cyclase (GenBank accession no. AB007002); LuS, lupeol synthases: OE-LU, O. europaea (GenBank accession no. AB025343); TO-LU, Taraxacum officinale (GenBank accession no. AB025345); βAS, β-amyrin synthases: AS PG-βAS, P. ginseng (GenBank accession no. AB009030); GG-βAS, G. glabra (GenBank accession no. AB037203); PS-βAS, P. sativum (GenBank accession no. AB034802); CS, cycloartenol synthases: AS-CS, A. strigosa AsCS1 (GenBank accession no. AJ311790); OS-CS, O. sativa (GenBank accession no. AF169966); LC-CS, Luffa cylindrica (GenBank accession no. AB033334); GG-CS, G. glabra (GenBank accession no. AB025968); PS-CS, P. sativum (GenBank accession no. D89619); PG-CS, P. ginseng (GenBank accession no. AB009029); OE-CS, O. europaea (GenBank accession no. AB025344). The phylogenetic tree was constructed by using the unweighted pair group method with arithmetic mean (UPGMA) method as implemented in the neighbor program of the phylip package (Version 3.5c; ref. 29). Amino acid distances were calculated by using the Dayhoff PAM matrix method of the protdist program of phylip. The numbers indicate the numbers of bootstrap replications (of 500) in which the given branching was observed. The protein parsimony method (the protpars program of phylip) produced trees with essentially identical topologies.