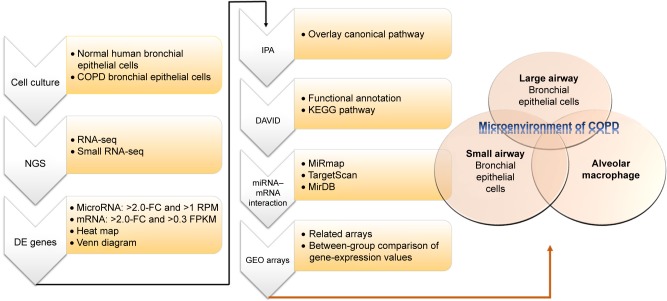
Figure 1.
Flowchart of study design.
Notes: In order to investigate the roles of microRNA–mRNA interactions in the microenvironment of COPD, we used normal human bronchial epithelial cells and COPD bronchial epithelial cells for next-generation sequencing (NGS). Then, we analyzed the NGS data of with several bioinformatic tools, including MiRmap, Ingenuity Pathway Analysis (IPA), the Database for Annotation, Visualization, and Integrated Discovery (DAVID), MirDB, TargetScan, and the Gene Expression Omnibus (GEO) database.
Abbreviations: DE, differentially expressed; FC, fold change; RPM, reads per million; FPKM, fragments per kilobase of transcript per million mapped reads; KEGG, Kyoto Encyclopedia of Genes and Genomes.